Genetics of Transformed Follicular Lymphoma
Abstract
1. Introduction
2. Definition of FL Transformation
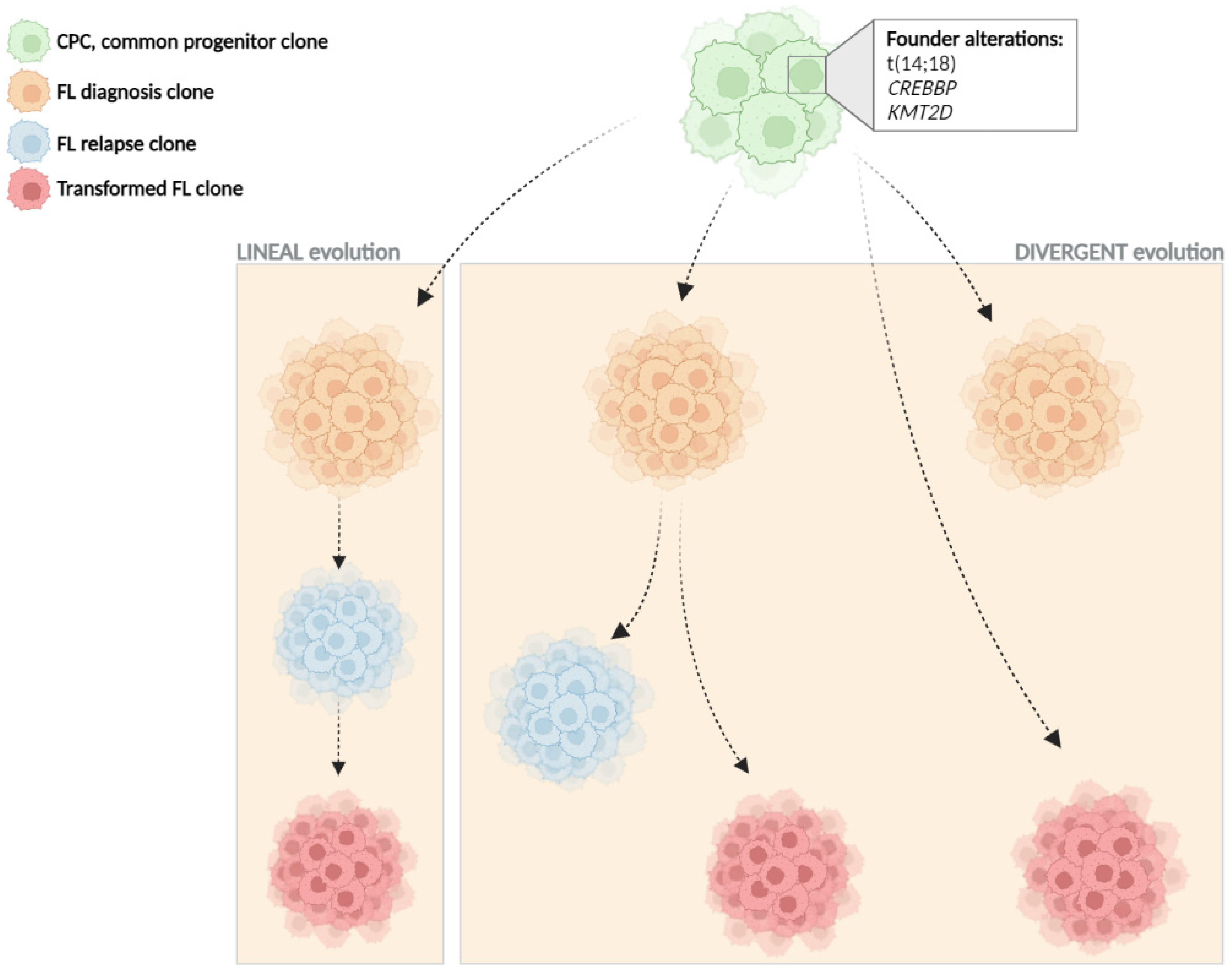
3. Clonal Evolution
4. Cell of Origin and Pathogenesis of FL Transformation
| Category | Variable | Biological Effect | Effect on Transformation |
|---|---|---|---|
| IHQ and microenvironment | IRF4 expression | - | Increased at HT [27] |
| MYC expression | - | Increased at HT [65] | |
| FOXP1 expression | - | Increased at HT [66] | |
| Genomic variants | TP53 mutation and deletion | Cell cycle | Increased at HT [28,39,41,42,55] |
| B2microglobulin mutation and deletion | Immune surveillance | Increased at HT [28,42] | |
| FAS mutation and deletion | Apoptosis | Enriched in transformed cases [42] | |
| MYC mutation and translocation | Cell cycle | Increased at HT [27,42] | |
| CCND3 mutation | Cell cycle, JAK-STAT signalling | Increased at HT [28,39] | |
| EBF1 mutation | B-cell development | Increased at HT [28,41] | |
| GNA13 mutation | NF-kB/BCR signalling | Increased at HT [28] | |
| P2RY8 mutation | B-cell migration | Increased at HT [28] | |
| S1PR2 mutation | Proliferation | Increased at HT [28] | |
| CD58 mutation | Immune surveillance | Increased at HT [42] | |
| MYD88 mutation | NF-kB/BCR signalling | Increased at HT, ABC-HT related [28,39,41,55] | |
| CD79B mutation | NF-kB/BCR signalling | Increased at HT, ABC-HT related [28,39,55] | |
| BCL10 mutation | NF-kB/BCR signalling | Increased at HT, ABC-HT related [28,55] | |
| CDKN2A/B deletion | Cell cycle | Increased at HT [28,41,42,54,57,61] | |
| BCL6 translocation | B-cell differentiation | Increased at HT [27,67] | |
| 2p16 (REL) amplification | NF-kB/BCR signalling | Increased at HT, GCB-HT related [35,42,54,60,61] | |
| 3q27.3-q28 (BCL6) gains | B-cell differentiation | Increased at HT [35,42,54,60] | |
| Chromosomes 2, 5 and 11 gains | - | Increased at HT [35,54] | |
| Genomic complexity -copy-number changes- | - | Increased at HT [28,41,42,53,54,55,56] | |
| Genetic complexity -mutations- | - | Increased at HT [28,39,41,42,55,68] |
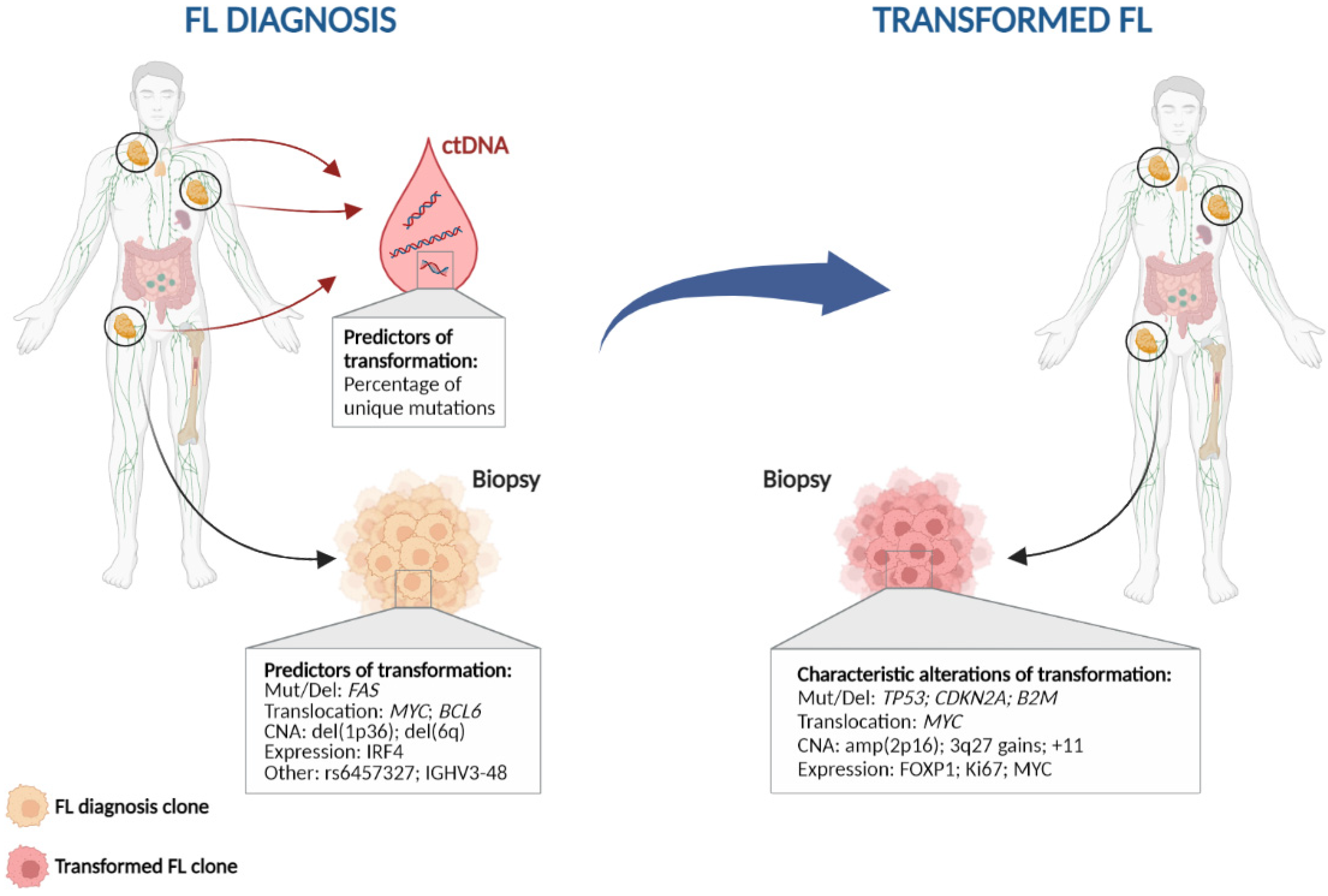
5. Can We Predict Transformation at Diagnosis?
5.1. Clinical, Biological and Immunohistochemical Factors
5.2. Genetic Aberrations
| Category | Variable | Effect on Transformation |
|---|---|---|
| Clinical | High FLIPI (≥3) | Higher risk of HT [18,19,20,21,22,32] |
| IHQ and microenvironment | FL Grade 3A | Higher risk of HT (controversial) [27,32] |
| High IRF4 expression | Higher risk of HT [27] | |
| High levels of lymphoma-associated macrophages | Shorter time to HT [106] | |
| High density of CD21 Follicular dendritic cells | Shorter time to HT, absent at HT [106] | |
| High levels of CD4+, CD8+, CD57+, PD1+, and FOXP3+ | Higher risk of HT [106] | |
| Follicular pattern of FOXP3+ T-cells | Higher risk of HT [107] | |
| Low tumour distance to blood vessels | Higher risk of HT [108] | |
| Genomic variants | 1p36, 6q deletions | Higher risk of HT [94,97] |
| BCL6, MYC translocations | Higher risk of HT [27,42,67] | |
| 16p CNN-LOH | Higher risk of HT [53] | |
| IGHV3-48 gene usage | Higher risk of HT [36] | |
| SNP rs6457327 (6p region) | Higher risk of HT [109] | |
| Circulating tumour DNA mutations | Higher risk of HT [46] |
5.3. Tumour Microenvironment
5.4. Liquid Biopsy
6. Discussion and Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Sant, M.; Allemani, C.; Tereanu, C.; De, A.R.; Capocaccia, R.; Visser, O.; Marcos-Gragera, R.; Maynadié, M.; Simonetti, A.; Lutz, J.M.; et al. Incidence of hematologic malignancies in Europe by morphologic subtype: Results of the HAEMACARE project. Blood 2010, 116, 3724–3734. [Google Scholar] [CrossRef] [PubMed]
- Kridel, R.; Sehn, L.H.; Gascoyne, R.D. Pathogenesis of follicular lymphoma. J. Clin. Investig. 2012, 122, 3424–3431. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Roulland, S.; Gloghini, A.; Younes, A.; von Keudell, G.; López-Guillermo, A.; Fitzgibbon, J. Follicular lymphoma. Nat. Rev. Dis. Primers 2019, 5, 83. [Google Scholar] [CrossRef]
- Bastos-Oreiro, M.; Muntañola, A.; Panizo, C.; Gonzalez-Barca, E.; de Villambrosia, S.G.; Córdoba, R.; López, J.L.B.; González-Sierra, P.; Terol, M.J.; Gutierrez, A.; et al. RELINF: Prospective epidemiological registry of lymphoid neoplasms in Spain. A project from the GELTAMO group. Ann. Hematol. 2020, 99, 799–808. [Google Scholar] [CrossRef]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A Report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Hiddemann, W.; Kneba, M.; Dreyling, M.; Schmitz, N.; Lengfelder, E.; Schmits, R.; Reiser, M.; Metzner, B.; Harder, H.; Hegewisch-Becker, S.; et al. Frontline therapy with rituximab added to the combination of cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) significantly improves the outcome for patients with advanced-stage follicular lymphoma compared with therapy with CHOP alone: Results of a prospective randomized study of the German Low-Grade Lymphoma Study Group. Blood 2005, 106, 3725–3732. [Google Scholar]
- Marcus, R.; Imrie, K.; Solal-Celigny, P.; Catalano, J.V.; Dmoszynska, A.; Raposo, J.C.; Offner, F.C.; Gomez-Codina, J.; Belch, A.; Cunningham, D.; et al. Phase III study of R-CVP compared with cyclophosphamide, vincristine, and prednisone alone in patients with previously untreated advanced follicular lymphoma. J. Clin. Oncol. 2008, 26, 4579–4586. [Google Scholar] [CrossRef]
- Salles, G.; Seymour, J.F.; Offner, F.; Lopez-Guillermo, A.; Belada, D.; Xerri, L.; Feugier, P.; Bouabdallah, R.; Catalano, J.V.; Brice, P.; et al. Rituximab maintenance for 2 years in patients with high tumour burden follicular lymphoma responding to rituximab plus chemotherapy (PRIMA): A phase 3, randomised controlled trial. Lancet 2011, 377, 42–51. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J.W. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; IARC Press: Lyon, France, 2008. [Google Scholar]
- Casulo, C.; Byrtek, M.; Dawson, K.L.; Zhou, X.; Farber, C.M.; Flowers, C.R.; Hainsworth, J.D.; Maurer, M.J.; Cerhan, J.R.; Link, B.K.; et al. Early Relapse of Follicular Lymphoma After Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone Defines Patients at High Risk for Death: An Analysis From the National LymphoCare Study. J. Clin. Oncol. 2015, 33, 2516–2522. [Google Scholar] [CrossRef] [PubMed]
- Sorigue, M.; Mercadal, S.; Alonso, S.; Fernández-Álvarez, R.; García, O.; Moreno, M.; Pomares, H.; Alcoceba, M.; González-García, E.; Motlló, C.; et al. Refractoriness to immunochemotherapy in follicular lymphoma: Predictive factors and outcome. Hematol. Oncol. 2017, 35, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Magnano, L.; Alonso-Alvarez, S.; Alcoceba, M.; Rivas-Delgado, A.; Muntañola, A.; Nadeu, F.; Setoain, X.; Rodríguez, S.; Andrade-Campos, M.; Espinosa-Lara, N.; et al. Life expectancy of follicular lymphoma patients in complete response at 30 months is similar to that of the Spanish general population. Br. J. Haematol. 2019, 185, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Federico, M.; Caballero, B.; Marcheselli, L.; Tarantino, V.; Manni, M.; Sarkozy, C.; Alonso-Álvarez, S.; Wondergem, M.; Cartron, G.; Lopez-Guillermo, A.; et al. Rituximab and the risk of transformation of follicular lymphoma: A retrospective pooled analysis. Lancet Haematol. 2018, 5, e359–e367. [Google Scholar] [CrossRef]
- Bastion, Y.; Sebban, C.; Berger, F.; Felman, P.; Salles, G.; Dumontet, C.; Bryon, P.A.; Coiffier, B. Incidence, predictive factors, and outcome of lymphoma transformation in follicular lymphoma patients. J. Clin. Oncol. 1997, 15, 1587–1594. [Google Scholar] [CrossRef]
- Montoto, S.; Davies, A.J.; Matthews, J.; Calaminici, M.; Norton, A.J.; Amess, J.; Vinnicombe, S.; Waters, R.; Rohatiner, A.Z.; Lister, T.A. Risk and clinical implications of transformation of follicular lymphoma to diffuse large B-cell lymphoma. J. Clin. Oncol. 2007, 25, 2426–2433. [Google Scholar] [CrossRef]
- Al-Tourah, A.J.; Gill, K.K.; Chhanabhai, M.; Hoskins, P.J.; Klasa, R.J.; Savage, K.J.; Sehn, L.H.; Shenkier, T.N.; Gascoyne, R.D.; Connors, J.M. Population-based analysis of incidence and outcome of transformed non-Hodgkin’s lymphoma. J. Clin. Oncol. 2008, 26, 5165–5169. [Google Scholar] [CrossRef]
- Link, B.K.; Maurer, M.J.; Nowakowski, G.S.; Ansell, S.M.; Macon, W.R.; Syrbu, S.I.; Slager, S.L.; Thompson, C.A.; Inwards, D.J.; Johnston, P.B.; et al. Rates and outcomes of follicular lymphoma transformation in the immunochemotherapy era: A report from the University of Iowa/MayoClinic Specialized Program of Research Excellence Molecular Epidemiology Resource. J. Clin. Oncol. 2013, 31, 3272–3278. [Google Scholar] [CrossRef]
- Wagner-Johnston, N.D.; Link, B.K.; Byrtek, M.; Dawson, K.L.; Hainsworth, J.; Flowers, C.R.; Friedberg, J.W.; Bartlett, N.L. Outcomes of transformed follicular lymphoma in the modern era: A report from the National LymphoCare Study (NLCS). Blood 2015, 126, 851–857. [Google Scholar] [CrossRef]
- Sarkozy, C.; Trneny, M.; Xerri, L.; Wickham, N.; Feugier, P.; Leppa, S.; Brice, P.; Soubeyran, P.; Gomes Da Silva, M.; Mounier, C.; et al. Risk Factors and Outcomes for Patients With Follicular Lymphoma Who Had Histologic Transformation After Response to First-Line Immunochemotherapy in the PRIMA Trial. J. Clin. Oncol. 2016, 34, 2575–2582. [Google Scholar] [CrossRef]
- Alonso-Álvarez, S.; Magnano, L.; Alcoceba, M.; Andrade-Campos, M.; Espinosa-Lara, N.; Rodríguez, G.; Mercadal, S.; Carro, I.; Sancho, J.M.; Moreno, M.; et al. Risk of, and survival following, histological transformation in follicular lymphoma in the rituximab era. A retrospective multicentre study by the Spanish GELTAMO group. Br. J. Haematol. 2017, 178, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Casulo, C.; Burack, W.R.; Friedberg, J.W. Transformed follicular non-Hodgkin lymphoma. Blood 2015, 125, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Kridel, R.; Sehn, L.H.; Gascoyne, R.D. Can histologic transformation of follicular lymphoma be predicted and prevented? Blood 2017, 130, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Lossos, I.S.; Gascoyne, R.D. Transformation of follicular lymphoma. Best Pract. Res. Clin. Haematol. 2011, 24, 147–163. [Google Scholar] [CrossRef]
- Ouansafi, I.; He, B.; Fraser, C.; Nie, K.; Mathew, S.; Bhanji, R.; Hoda, R.; Arabadjief, M.; Knowles, D.; Cerutti, A.; et al. Transformation of follicular lymphoma to plasmablastic lymphoma with c-myc gene rearrangement. Am. J. Clin. Pathol. 2010, 134, 972–981. [Google Scholar] [CrossRef]
- Kridel, R.; Mottok, A.; Farinha, P.; Ben-Neriah, S.; Ennishi, D.; Zheng, Y.; Chavez, E.A.; Shulha, H.P.; Tan, K.; Chan, F.C.; et al. Cell of origin of transformed follicular lymphoma. Blood 2015, 126, 2118–2127. [Google Scholar] [CrossRef]
- Kridel, R.; Chan, F.C.; Mottok, A.; Boyle, M.; Farinha, P.; Tan, K.; Meissner, B.; Bashashati, A.; McPherson, A.; Roth, A.; et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016, 13, e1002197. [Google Scholar] [CrossRef]
- Salles, G.; Coiffier, B. Histologic transformation in follicular lymphoma. Ann. Oncol. 1998, 9, 803–805. [Google Scholar] [CrossRef]
- Araf, S.; Wang, J.; Korfi, K.; Pangault, C.; Kotsiou, E.; Rio-Machin, A.; Rahim, T.; Heward, J.; Clear, A.; Iqbal, S.; et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia 2018, 32, 1261–1265. [Google Scholar] [CrossRef]
- Bodet-Milin, C.; Kraeber-Bodere, F.; Moreau, P.; Campion, L.; Dupas, B.; Le, G.S. Investigation of FDG-PET/CT imaging to guide biopsies in the detection of histological transformation of indolent lymphoma. Haematologica 2008, 93, 471–472. [Google Scholar] [CrossRef]
- Gine, E.; Montoto, S.; Bosch, F.; Arenillas, L.; Mercadal, S.; Villamor, N.; Martinez, A.; Colomo, L.; Campo, E.; Montserrat, E.; et al. The Follicular Lymphoma International Prognostic Index (FLIPI) and the histological subtype are the most important factors to predict histological transformation in follicular lymphoma. Ann. Oncol. 2006, 17, 1539–1545. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, R.D. The pathology of transformation of indolent B cell lymphomas. Hematol. Oncol. 2015, 33 (Suppl. 1), 75–79. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Spina, V.; Deambrogi, C.; Rasi, S.; Laurenti, L.; Stamatopoulos, K.; Arcaini, L.; Lucioni, M.; Rocque, G.B.; Xu-Monette, Z.Y.; et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood 2011, 117, 3391–3401. [Google Scholar] [CrossRef]
- Eide, M.B.; Liestol, K.; Lingjaerde, O.C.; Hystad, M.E.; Kresse, S.H.; Meza-Zepeda, L.; Myklebost, O.; Troen, G.; Aamot, H.V.; Holte, H.; et al. Genomic alterations reveal potential for higher grade transformation in follicular lymphoma and confirm parallel evolution of tumor cell clones. Blood 2010, 116, 1489–1497. [Google Scholar] [CrossRef]
- García-Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Maldonado, R.; Antón, A.; et al. Immunoglobulin gene rearrangement IGHV3–48 is a predictive marker of histological transformation into aggressive lymphoma in follicular lymphomas. Blood Cancer J. 2019, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Fitzgibbon, J.; Iqbal, S.; Davies, A.; O’Shea, D.; Carlotti, E.; Chaplin, T.; Matthews, J.; Raghavan, M.; Norton, A.; Lister, T.A.; et al. Genome-wide detection of recurring sites of uniparental disomy in follicular and transformed follicular lymphoma. Leukemia 2007, 21, 514–1520. [Google Scholar] [CrossRef]
- Johnson, N.A.; Al-Tourah, A.; Brown, C.J.; Connors, J.M.; Gascoyne, R.D.; Horsman, D.E. Prognostic significance of secondary cytogenetic alterations in follicular lymphomas. Genes Chromosom. Cancer 2008, 47, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, M.I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Antón, A.; Maldonado, R.; et al. Molecular study of the clonal evolution of follicular lymphoma to aggressive lymphoma. A single center experience. Haematologica 2018, 103, 15. [Google Scholar]
- González-Rincón, J.; Méndez, M.; Gómez, S.; García, J.F.; Martín, P.; Bellas, C.; Pedrosa, L.; Rodríguez-Pinilla, S.M.; Camacho, F.I.; Quero, C.; et al. Unraveling transformation of follicular lymphoma to diffuse large B-cell lymphoma. PLoS ONE 2019, 14, e0212813. [Google Scholar] [CrossRef]
- Okosun, J.; Bodor, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Khiabanian, H.; Fangazio, M.; Vasishtha, M.; Messina, M.; Holmes, A.B.; Ouillette, P.; Trifonov, V.; Rossi, D.; Tabbo, F.; et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014, 6, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, G.; Khiabanian, H.; Holmes, A.B.; Wang, J.; Messina, M.; Mullighan, C.G.; Pasqualucci, L.; Rabadan, R.; Dalla-Favera, R. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J. Exp. Med. 2013, 210, 2273–2288. [Google Scholar] [CrossRef] [PubMed]
- Carlotti, E.; Wrench, D.; Matthews, J.; Iqbal, S.; Davies, A.; Norton, A.; Hart, J.; Lai, R.; Montoto, S.; Gribben, J.G.; et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma may occur by divergent evolution from a common progenitor cell or by direct evolution from the follicular lymphoma clone. Blood 2009, 113, 3553–3557. [Google Scholar] [CrossRef]
- Weigert, O.; Kopp, N.; Lane, A.A.; Yoda, A.; Dahlberg, S.E.; Neuberg, D.; Bahar, A.Y.; Chapuy, B.; Kutok, J.L.; Longtine, J.A.; et al. Molecular ontogeny of donor-derived follicular lymphomas occurring after hematopoietic cell transplantation. Cancer Discov. 2012, 2, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Scherer, F.; Kurtz, D.M.; Newman, A.M.; Stehr, H.; Craig, A.F.; Esfahani, M.S.; Lovejoy, A.F.; Chabon, J.J.; Klass, D.M.; Liu, C.L.; et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 2016, 8, 364ra155. [Google Scholar] [CrossRef]
- Roulland, S.; Kelly, R.S.; Morgado, E.; Sungalee, S.; Solal-Celigny, P.; Colombat, P.; Jouve, N.; Palli, D.; Pala, V.; Tumino, R.; et al. t(14;18) Translocation: A predictive blood biomarker for follicular lymphoma. J. Clin. Oncol. 2014, 32, 1347–1355. [Google Scholar] [CrossRef]
- Davies, A.J.; Rosenwald, A.; Wright, G.; Lee, A.; Last, K.W.; Weisenburger, D.D.; Chan, W.C.; Delabie, J.; Braziel, R.M.; Campo, E.; et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma proceeds by distinct oncogenic mechanisms. Br. J. Haematol. 2007, 136, 286–293. [Google Scholar] [CrossRef]
- Maeshima, A.M.; Taniguchi, H.; Toyoda, K.; Yamauchi, N.; Makita, S.; Fukuhara, S.; Munakata, W.; Maruyama, D.; Kobayashi, Y.; Tobinai, K. Clinicopathological features of histological transformation from extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue to diffuse large B-cell lymphoma: An analysis of 467 patients. Br. J. Haematol. 2016, 174, 923–931. [Google Scholar] [CrossRef]
- Zanwar, S.; Abeykoon, J.P.; Durot, E.; King, R.; Perez Burbano, G.E.; Kumar, S.; Gertz, M.A.; Quinquenel, A.; Delmer, A.; Gonsalves, W.; et al. Impact of MYD88(L265P) mutation status on histological transformation of Waldenström Macroglobulinemia. Am. J. Hematol. 2020, 95, 274–281. [Google Scholar] [CrossRef]
- Abrisqueta, P.; Delgado, J.; Alcoceba, M.; Oliveira, A.C.; Loscertales, J.; Hernández-Rivas, J.A.; Ferrà, C.; Cordoba, R.; Yáñez, L.; Medina, A.; et al. Clinical outcome and prognostic factors of patients with Richter syndrome: Real-world study of the Spanish Chronic Lymphocytic Leukemia Study Group (GELLC). Br. J. Haematol. 2020, 190, 854–863. [Google Scholar] [CrossRef]
- Bastidas-Mora, G.; Beà, S.; Navarro, A.; Gine, E.; Costa, D.; Delgado, J.; Baumann, T.; Magnano, L.; Rivas-Delgado, A.; Villamor, N.; et al. Clinico-biological features and outcome of patients with splenic marginal zone lymphoma with histological transformation. Br. J. Haematol. 2022, 196, 146–155. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, D.; O’Riain, C.; Gupta, M.; Waters, R.; Yang, Y.; Wrench, D.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. Regions of acquired uniparental disomy at diagnosis of follicular lymphoma are associated with both overall survival and risk of transformation. Blood 2009, 113, 2298–2301. [Google Scholar] [CrossRef] [PubMed]
- Bouska, A.; McKeithan, T.W.; Deffenbacher, K.E.; Lachel, C.; Wright, G.W.; Iqbal, J.; Smith, L.M.; Zhang, W.; Kucuk, C.; Rinaldi, A.; et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood 2014, 123, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Bouska, A.; Zhang, W.; Gong, Q.; Iqbal, J.; Scuto, A.; Vose, J.; Ludvigsen, M.; Fu, K.; Weisenburger, D.D.; Greiner, T.C.; et al. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia 2017, 31, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Li, H.; Braziel, R.M.; Passerini, V.; Rimsza, L.M.; Hsi, E.D.; Leonard, J.P.; Smith, S.M.; Kridel, R.; Press, O.; et al. Genomic alterations important for the prognosis in patients with follicular lymphoma treated in SWOG study S0016. Blood 2019, 133, 81–93. [Google Scholar] [CrossRef]
- Elenitoba-Johnson, K.S.; Gascoyne, R.D.; Lim, M.S.; Chhanabai, M.; Jaffe, E.S.; Raffeld, M. Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood 1998, 91, 4677–4685. [Google Scholar] [CrossRef]
- Alhejaily, A.; Day, A.G.; Feilotter, H.E.; Baetz, T.; Lebrun, D.P. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin. Cancer Res. 2014, 20, 1676–1686. [Google Scholar] [CrossRef]
- Martello, M.; Poletti, A.; Borsi, E.; Solli, V.; Dozza, L.; Barbato, S.; Zamagni, E.; Tacchetti, P.; Pantani, L.; Mancuso, K.; et al. Clonal and subclonal TP53 molecular impairment is associated with prognosis and progression in multiple myeloma. Blood Cancer J. 2022, 12, 15. [Google Scholar] [CrossRef]
- Martinez-Climent, J.A.; Alizadeh, A.A.; Segraves, R.; Blesa, D.; Rubio-Moscardo, F.; Albertson, D.G.; Garcia-Conde, J.; Dyer, M.J.; Levy, R.; Pinkel, D.; et al. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood 2003, 101, 3109–3117. [Google Scholar] [CrossRef]
- Kwiecinska, A.; Ichimura, K.; Berglund, M.; Dinets, A.; Sulaiman, L.; Collins, V.P.; Larsson, C.; Porwit, A.; Lagercrantz, S.B. Amplification of 2p as a genomic marker for transformation in lymphoma. Genes Chromosom. Cancer 2014, 53, 750–768. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Chigrinova, E.; Rinaldi, A.; Kwee, I.; Rossi, D.; Rancoita, P.M.; Strefford, J.C.; Oscier, D.; Stamatopoulos, K.; Papadaki, T.; Berger, F.; et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood 2013, 122, 2673–2682. [Google Scholar] [CrossRef]
- Aukema, S.M.; van Pel, R.; Nagel, I.; Bens, S.; Siebert, R.; Rosati, S.; van den Berg, E.; Bosga-Bouwer, A.G.; Kibbelaar, R.E.; Hoogendoorn, M.; et al. MYC expression and translocation analyses in low-grade and transformed follicular lymphoma. Histopathology 2017, 71, 960–971. [Google Scholar] [CrossRef]
- Musilova, K.; Devan, J.; Cerna, K.; Seda, V.; Pavlasova, G.; Sharma, S.; Oppelt, J.; Pytlik, R.; Prochazka, V.; Prouzova, Z.; et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood 2018, 132, 2389–2400. [Google Scholar] [CrossRef]
- Akasaka, T.; Lossos, I.S.; Levy, R. BCL6 gene translocation in follicular lymphoma: A harbinger of eventual transformation to diffuse aggressive lymphoma. Blood 2003, 102, 1443–1448. [Google Scholar] [CrossRef]
- García Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Maldonado, R.; Antón, A.; et al. Genetic complexity impacts the clinical outcome of follicular lymphoma patients. Blood Cancer J. 2021, 11, 11. [Google Scholar] [CrossRef]
- Mozas, P.; Rivero, A.; Rivas-Delgado, A.; Correa, J.G.; Condom, M.; Nadeu, F.; Giné, E.; Delgado, J.; Villamor, N.; Campo, E.; et al. Prognostic ability of five clinical risk scores in follicular lymphoma: A single-center evaluation. Hematol. Oncol. 2021, 39, 639–649. [Google Scholar] [CrossRef]
- Alonso-Álvarez, S.; Manni, M.; Montoto, S.; Sarkozy, C.; Morschhauser, F.; Wondergem, M.J.; Guarini, A.; Magnano, L.; Alcoceba, M.; Chamuleau, M.; et al. Primary refractory follicular lymphoma: A poor outcome entity with high risk of transformation to aggressive B cell lymphoma. Eur. J. Cancer 2021, 157, 132–139. [Google Scholar] [CrossRef]
- Sarkozy, C.; Maurer, M.J.; Link, B.K.; Ghesquieres, H.; Nicolas, E.; Thompson, C.A.; Traverse-Glehen, A.; Feldman, A.L.; Allmer, C.; Slager, S.L.; et al. Cause of Death in Follicular Lymphoma in the First Decade of the Rituximab Era: A Pooled Analysis of French and US Cohorts. J. Clin. Oncol. 2019, 37, 144–152. [Google Scholar] [CrossRef]
- Naresh, K.N. MUM1 expression dichotomises follicular lymphoma into predominantly, MUM1-negative low-grade and MUM1-positive high-grade subtypes. Haematologica 2007, 92, 267–268. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koch, K.; Hoster, E.; Ziepert, M.; Unterhalt, M.; Ott, G.; Rosenwald, A.; Hansmann, M.L.; Bernd, W.; Stein, H.; Pöschel, V.; et al. Clinical, pathological and genetic features of follicular lymphoma grade 3A: A joint analysis of the German low-grade and high-grade lymphoma study groups GLSG and DSHNHL. Ann. Oncol. 2016, 27, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Sweetenham, J.W.; Goldman, B.; LeBlanc, M.L.; Cook, J.R.; Tubbs, R.R.; Press, O.W.; Maloney, D.G.; Fisher, R.I.; Rimsza, L.M.; Braziel, R.M.; et al. Prognostic value of regulatory T cells, lymphoma-associated macrophages, and MUM-1 expression in follicular lymphoma treated before and after the introduction of monoclonal antibody therapy: A Southwest Oncology Group Study. Ann. Oncol. 2010, 21, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Xerri, L.; Bachy, E.; Fabiani, B.; Canioni, D.; Chassagne-Clement, C.; Dartigues-Cuilleres, P.; Charlotte, F.; Brousse, N.; Rousselet, M.C.; Foussard, C.; et al. Identification of MUM1 as a prognostic immunohistochemical marker in follicular lymphoma using computerized image analysis. Hum. Pathol. 2014, 45, 2085–2093. [Google Scholar] [CrossRef]
- Mottok, A.; Jurinovic, V.; Farinha, P.; Rosenwald, A.; Leich, E.; Ott, G.; Horn, H.; Klapper, W.; Boesl, M.; Hiddemann, W.; et al. FOXP1 expression is a prognostic biomarker in follicular lymphoma treated with rituximab and chemotherapy. Blood 2018, 131, 226–235. [Google Scholar] [CrossRef]
- Sohani, A.R.; Maurer, M.J.; Giri, S.; Pitcher, B.; Chadburn, A.; Said, J.W.; Bartlett, N.L.; Czuczman, M.S.; Martin, P.; Rosenbaum, C.A.; et al. Biomarkers for Risk Stratification in Patients With Previously Untreated Follicular Lymphoma Receiving Anti-CD20-based Biological Therapy. Am. J. Surg. Pathol. 2021, 45, 384–393. [Google Scholar] [CrossRef]
- Sagardoy, A.; Martinez-Ferrandis, J.I.; Roa, S.; Bunting, K.L.; Aznar, M.A.; Elemento, O.; Shaknovich, R.; Fontán, L.; Fresquet, V.; Perez-Roger, I.; et al. Downregulation of FOXP1 is required during germinal center B-cell function. Blood 2013, 121, 4311–4320. [Google Scholar] [CrossRef]
- Gascoyne, D.M.; Banham, A.H. The significance of FOXP1 in diffuse large B-cell lymphoma. Leuk. Lymphoma 2017, 58, 1037–1051. [Google Scholar] [CrossRef]
- Patzelt, T.; Keppler, S.J.; Gorka, O.; Thoene, S.; Wartewig, T.; Reth, M.; Förster, I.; Lang, R.; Buchner, M.; Ruland, J. Foxp1 controls mature B cell survival and the development of follicular and B-1 B cells. Proc. Natl. Acad. Sci. USA 2018, 115, 3120–3125. [Google Scholar] [CrossRef]
- Barrans, S.L.; Fenton, J.A.; Banham, A.; Owen, R.G.; Jack, A.S. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood 2004, 104, 2933–2935. [Google Scholar] [CrossRef]
- Noppe, S.M.; Heirman, C.; Bakkus, M.H.; Brissinck, J.; Schots, R.; Thielemans, K. The genetic variability of the VH genes in follicular lymphoma: The impact of the hypermutation mechanism. Br. J. Haematol. 1999, 107, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Berget, E.; Molven, A.; Lokeland, T.; Helgeland, L.; Vintermyr, O.K. IGHV gene usage and mutational status in follicular lymphoma: Correlations with prognosis and patient age. Leuk. Res. 2015, 39, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Cerri, M.; Capello, D.; Deambrogi, C.; Rossi, F.M.; Zucchetto, A.; De, P.L.; Cresta, S.; Rasi, S.; Spina, V.; et al. Biological and clinical risk factors of chronic lymphocytic leukaemia transformation to Richter syndrome. Br. J. Haematol. 2008, 142, 202–215. [Google Scholar] [CrossRef]
- O’Shea, D.; O’Riain, C.; Taylor, C.; Waters, R.; Carlotti, E.; Macdougall, F.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. The presence of TP53 mutation at diagnosis of follicular lymphoma identifies a high-risk group of patients with shortened time to disease progression and poorer overall survival. Blood 2008, 112, 3126–3129. [Google Scholar] [CrossRef] [PubMed]
- Pastore, A.; Jurinovic, V.; Kridel, R.; Hoster, E.; Staiger, A.M.; Szczepanowski, M.; Pott, C.; Kopp, N.; Murakami, M.; Horn, H.; et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: A retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015, 16, 1111–1122. [Google Scholar] [CrossRef]
- Jurinovic, V.; Kridel, R.; Staiger, A.M.; Szczepanowski, M.; Horn, H.; Dreyling, M.H.; Rosenwald, A.; Ott, G.; Klapper, W.; Zelenetz, A.D.; et al. Clinicogenetic risk models predict early progression of follicular lymphoma after first-line immunochemotherapy. Blood 2016, 128, 1112–1120. [Google Scholar] [CrossRef]
- Krysiak, K.; Gomez, F.; White, B.S.; Matlock, M.; Miller, C.A.; Trani, L.; Fronick, C.C.; Fulton, R.S.; Kreisel, F.; Cashen, A.F.; et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood 2017, 129, 473–483. [Google Scholar] [CrossRef]
- Miao, Y.; Hu, S.; Lu, X.; Li, S.; Wang, W.; Medeiros, L.J.; Lin, P. Double-hit follicular lymphoma with MYC and BCL2 translocations: A study of 7 cases with a review of literature. Hum. Pathol. 2016, 58, 72–77. [Google Scholar] [CrossRef]
- Chaudhary, S.; Brown, N.; Song, J.Y.; Yang, L.; Skrabek, P.; Nasr, M.R.; Wong, J.T.; Bedell, V.; Murata-Collins, J.; Kochan, L.; et al. Relative frequency and clinicopathologic characteristics of MYC-rearranged follicular lymphoma. Hum. Pathol. 2021, 114, 19–27. [Google Scholar] [CrossRef]
- Bussot, L.; Chevalier, S.; Cristante, J.; Grange, B.; Tesson, B.; Deteix-Santana, C.; Orsini-Piocelle, F.; Leyronnas, C.; Dupire, S.; Gressin, R.; et al. Adverse outcome in follicular lymphoma is associated with MYC rearrangements but not MYC extra copies. Br. J. Haematol. 2021, 194, 382–392. [Google Scholar] [CrossRef]
- Razzaghi, R.; Agarwal, S.; Kotlov, N.; Plotnikova, O.; Nomie, K.; Huang, D.W.; Wright, G.W.; Smith, G.A.; Li, M.; Takata, K.; et al. Compromised counterselection by FAS creates an aggressive subtype of germinal center lymphoma. J. Exp. Med. 2021, 218, e20201173. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Greiner, T.C.; Patel, K.; Dave, B.J.; Smith, L.; Ji, J.; Wright, G.; Sanger, W.G.; Pickering, D.L.; Jain, S.; et al. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia 2007, 21, 2332–2343. [Google Scholar] [CrossRef] [PubMed]
- Tilly, H.; Rossi, A.; Stamatoullas, A.; Lenormand, B.; Bigorgne, C.; Kunlin, A.; Monconduit, M.; Bastard, C. Prognostic value of chromosomal abnormalities in follicular lymphoma. Blood 1994, 84, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Viardot, A.; Möller, P.; Högel, J.; Werner, K.; Mechtersheimer, G.; Ho, A.D.; Ott, G.; Barth, T.F.; Siebert, R.; Gesk, S.; et al. Clinicopathologic correlations of genomic gains and losses in follicular lymphoma. J. Clin. Oncol. 2002, 20, 4523–4530. [Google Scholar] [CrossRef]
- Schwaenen, C.; Viardot, A.; Berger, H.; Barth, T.F.; Bentink, S.; Döhner, H.; Enz, M.; Feller, A.C.; Hansmann, M.L.; Hummel, M.; et al. Microarray-based genomic profiling reveals novel genomic aberrations in follicular lymphoma which associate with patient survival and gene expression status. Genes Chromosom. Cancer 2009, 48, 39–54. [Google Scholar] [CrossRef]
- Cheung, K.J.; Shah, S.P.; Steidl, C.; Johnson, N.; Relander, T.; Telenius, A.; Lai, B.; Murphy, K.P.; Lam, W.; Al-Tourah, A.J.; et al. Genome-wide profiling of follicular lymphoma by array comparative genomic hybridization reveals prognostically significant DNA copy number imbalances. Blood 2009, 113, 137–148. [Google Scholar] [CrossRef]
- Cheung, K.J.; Johnson, N.A.; Affleck, J.G.; Severson, T.; Steidl, C.; Ben-Neriah, S.; Schein, J.; Morin, R.D.; Moore, R.; Shah, S.P.; et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010, 70, 9166–9174. [Google Scholar] [CrossRef]
- Murphy, K.M.; Nelson, C.A.; Sedý, J.R. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat. Rev. Immunol. 2006, 6, 671–681. [Google Scholar] [CrossRef]
- Cai, G.; Freeman, G.J. The CD160, BTLA, LIGHT/HVEM pathway: A bidirectional switch regulating T-cell activation. Immunol. Rev. 2009, 229, 244–258. [Google Scholar] [CrossRef]
- Kotsiou, E.; Okosun, J.; Besley, C.; Iqbal, S.; Matthews, J.; Fitzgibbon, J.; Gribben, J.G.; Davies, J.K. TNFRSF14 aberrations in follicular lymphoma increase clinically significant allogeneic T-cell responses. Blood 2016, 128, 72–81. [Google Scholar] [CrossRef]
- Boice, M.; Salloum, D.; Mourcin, F.; Sanghvi, V.; Amin, R.; Oricchio, E.; Jiang, M.; Mottok, A.; Denis-Lagache, N.; Ciriello, G.; et al. Loss of the HVEM Tumor Suppressor in Lymphoma and Restoration by Modified CAR-T Cells. Cell 2016, 167, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Kihira, S.; Liu, C.L.; Nair, R.V.; Salari, R.; Gentles, A.J.; Irish, J.; Stehr, H.; Vicente-Dueñas, C.; Romero-Camarero, I.; et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc. Natl. Acad. Sci. USA 2015, 112, E1116–E1125. [Google Scholar] [CrossRef] [PubMed]
- Okosun, J.; Wolfson, R.L.; Wang, J.; Araf, S.; Wilkins, L.; Castellano, B.M.; Escudero-Ibarz, L.; Al Seraihi, A.F.; Richter, J.; Bernhart, S.H.; et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat. Genet. 2016, 48, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Bararia, D.; Hildebrand, J.A.; Stolz, S.; Haebe, S.; Alig, S.; Trevisani, C.P.; Osorio-Barrios, F.; Bartoschek, M.D.; Mentz, M.; Pastore, A.; et al. Cathepsin S Alterations Induce a Tumor-Promoting Immune Microenvironment in Follicular Lymphoma. Cell Rep. 2020, 31, 107522. [Google Scholar] [CrossRef] [PubMed]
- Blaker, Y.N.; Spetalen, S.; Brodtkorb, M.; Lingjaerde, O.C.; Beiske, K.; Ostenstad, B.; Sander, B.; Wahlin, B.E.; Melen, C.M.; Myklebust, J.H.; et al. The tumour microenvironment influences survival and time to transformation in follicular lymphoma in the rituximab era. Br. J. Haematol. 2016, 175, 102–114. [Google Scholar] [CrossRef]
- Farinha, P.; Al-Tourah, A.; Gill, K.; Klasa, R.; Connors, J.M.; Gascoyne, R.D. The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood 2010, 115, 289–295. [Google Scholar] [CrossRef]
- Farinha, P.; Kyle, A.H.; Minchinton, A.I.; Connors, J.M.; Karsan, A.; Gascoyne, R.D. Vascularization predicts overall survival and risk of transformation in follicular lymphoma. Haematologica 2010, 95, 2157–2160. [Google Scholar] [CrossRef]
- Berglund, M.; Enblad, G.; Thunberg, U. SNP rs6457327 is a predictor for overall survival in follicular lymphoma as well as survival after transformation. Blood 2011, 118, 4489. [Google Scholar] [CrossRef][Green Version]
- Gentles, A.J.; Alizadeh, A.A.; Lee, S.I.; Myklebust, J.H.; Shachaf, C.M.; Shahbaba, B.; Levy, R.; Koller, D.; Plevritis, S.K. A pluripotency signature predicts histologic transformation and influences survival in follicular lymphoma patients. Blood 2009, 114, 3158–3166. [Google Scholar] [CrossRef]
- Brodtkorb, M.; Lingjaerde, O.C.; Huse, K.; Troen, G.; Hystad, M.; Hilden, V.I.; Myklebust, J.H.; Leich, E.; Rosenwald, A.; Delabie, J.; et al. Whole-genome integrative analysis reveals expression signatures predicting transformation in follicular lymphoma. Blood 2014, 123, 1051–1054. [Google Scholar] [CrossRef]
- Steen, C.B.; Leich, E.; Myklebust, J.H.; Lockmer, S.; Wise, J.F.; Wahlin, B.E.; Østenstad, B.; Liestøl, K.; Kimby, E.; Rosenwald, A.; et al. A clinico-molecular predictor identifies follicular lymphoma patients at risk of early transformation after first-line immunotherapy. Haematologica 2019, 104, e460–e464. [Google Scholar] [CrossRef] [PubMed]
- Farinha, P.; Masoudi, H.; Skinnider, B.F.; Shumansky, K.; Spinelli, J.J.; Gill, K.; Klasa, R.; Voss, N.; Connors, J.M.; Gascoyne, R.D. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood 2005, 106, 2169–2174. [Google Scholar] [CrossRef] [PubMed]
- Alvaro, T.; Lejeune, M.; Camacho, F.I.; Salvado, M.T.; Sanchez, L.; Garcia, J.F.; Lopez, C.; Jaen, J.; Bosch, R.; Pons, L.E.; et al. The presence of STAT1-positive tumor-associated macrophages and their relation to outcome in patients with follicular lymphoma. Haematologica 2006, 91, 1605–1612. [Google Scholar] [PubMed]
- Taskinen, M.; Karjalainen-Lindsberg, M.L.; Nyman, H.; Eerola, L.M.; Leppa, S. A high tumor-associated macrophage content predicts favorable outcome in follicular lymphoma patients treated with rituximab and cyclophosphamide-doxorubicin-vincristine-prednisone. Clin. Cancer Res. 2007, 13, 5784–5789. [Google Scholar] [CrossRef]
- Canioni, D.; Salles, G.; Mounier, N.; Brousse, N.; Keuppens, M.; Morchhauser, F.; Lamy, T.; Sonet, A.; Rousselet, M.C.; Foussard, C.; et al. High numbers of tumor-associated macrophages have an adverse prognostic value that can be circumvented by rituximab in patients with follicular lymphoma enrolled onto the GELA-GOELAMS FL-2000 trial. J. Clin. Oncol. 2008, 26, 440–446. [Google Scholar] [CrossRef]
- Richendollar, B.G.; Pohlman, B.; Elson, P.; Hsi, E.D. Follicular programmed death 1-positive lymphocytes in the tumor microenvironment are an independent prognostic factor in follicular lymphoma. Hum. Pathol. 2011, 42, 552–557. [Google Scholar] [CrossRef]
- Leidi, M.; Gotti, E.; Bologna, L.; Miranda, E.; Rimoldi, M.; Sica, A.; Roncalli, M.; Palumbo, G.A.; Introna, M.; Golay, J. M2 macrophages phagocytose rituximab-opsonized leukemic targets more efficiently than m1 cells in vitro. J. Immunol. 2009, 182, 4415–4422. [Google Scholar] [CrossRef]
- de Jong, D.; Koster, A.; Hagenbeek, A.; Raemaekers, J.; Veldhuizen, D.; Heisterkamp, S.; de Boer, J.P.; van Glabbeke, M. Impact of the tumor microenvironment on prognosis in follicular lymphoma is dependent on specific treatment protocols. Haematologica 2009, 94, 70–77. [Google Scholar] [CrossRef]
- Smeltzer, J.P.; Jones, J.M.; Ziesmer, S.C.; Grote, D.M.; Xiu, B.; Ristow, K.M.; Yang, Z.Z.; Nowakowski, G.S.; Feldman, A.L.; Cerhan, J.R.; et al. Pattern of CD14+ follicular dendritic cells and PD1+ T cells independently predicts time to transformation in follicular lymphoma. Clin. Cancer Res. 2014, 20, 2862–2872. [Google Scholar] [CrossRef]
- Alvaro, T.; Lejeune, M.; Salvadó, M.T.; Lopez, C.; Jaén, J.; Bosch, R.; Pons, L.E. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients. J. Clin. Oncol. 2006, 24, 5350–5357. [Google Scholar] [CrossRef]
- Wahlin, B.E.; Aggarwal, M.; Montes-Moreno, S.; Gonzalez, L.F.; Roncador, G.; Sanchez-Verde, L.; Christensson, B.; Sander, B.; Kimby, E. A unifying microenvironment model in follicular lymphoma: Outcome is predicted by programmed death-1--positive, regulatory, cytotoxic, and helper T cells and macrophages. Clin. Cancer Res. 2010, 16, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Laurent, C.; Müller, S.; Do, C.; Al-Saati, T.; Allart, S.; Larocca, L.M.; Hohaus, S.; Duchez, S.; Quillet-Mary, A.; Laurent, G.; et al. Distribution, function, and prognostic value of cytotoxic T lymphocytes in follicular lymphoma: A 3-D tissue-imaging study. Blood 2011, 118, 5371–5379. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.; Lauridsen, K.L.; Plesner, T.L.; Monrad, I.; Honoré, B.; Hamilton-Dutoit, S.; D’Amore, F.; Ludvigsen, M. High intratumoral expression of vimentin predicts histological transformation in patients with follicular lymphoma. Blood Cancer J. 2019, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Kridel, R.; Xerri, L.; Gelas-Dore, B.; Tan, K.; Feugier, P.; Vawda, A.; Canioni, D.; Farinha, P.; Boussetta, S.; Moccia, A.A.; et al. The Prognostic Impact of CD163-Positive Macrophages in Follicular Lymphoma: A Study from the BC Cancer Agency and the Lymphoma Study Association. Clin. Cancer Res. 2015, 21, 3428–3435. [Google Scholar] [CrossRef]
- Menter, T.; Tzankov, A.; Zucca, E.; Kimby, E.; Hultdin, M.; Sundström, C.; Beiske, K.; Cogliatti, S.; Banz, Y.; Cathomas, G.; et al. Prognostic implications of the microenvironment for follicular lymphoma under immunomodulation therapy. Br. J. Haematol. 2020, 189, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Taskinen, M.; Karjalainen-Lindsberg, M.L.; Leppa, S. Prognostic influence of tumor-infiltrating mast cells in patients with follicular lymphoma treated with rituximab and CHOP. Blood 2008, 111, 4664–4667. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.; Neelapu, S.S. Anti-PD-1 antibodies for the treatment of B-cell lymphoma: Importance of PD-1(+) T-cell subsets. Oncoimmunology 2014, 3, e28101. [Google Scholar] [CrossRef]
- Skibola, C.F.; Bracci, P.M.; Halperin, E.; Conde, L.; Craig, D.W.; Agana, L.; Iyadurai, K.; Becker, N.; Brooks-Wilson, A.; Curry, J.D.; et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat. Genet. 2009, 41, 873–875. [Google Scholar] [CrossRef]
- Conde, L.; Halperin, E.; Akers, N.K.; Brown, K.M.; Smedby, K.E.; Rothman, N.; Nieters, A.; Slager, S.L.; Brooks-Wilson, A.; Agana, L.; et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat. Genet. 2010, 42, 661–664. [Google Scholar] [CrossRef]
- Wang, S.S.; Abdou, A.M.; Morton, L.M.; Thomas, R.; Cerhan, J.R.; Gao, X.; Cozen, W.; Rothman, N.; Davis, S.; Severson, R.K.; et al. Human leukocyte antigen class I and II alleles in non-Hodgkin lymphoma etiology. Blood 2010, 115, 4820–4823. [Google Scholar] [CrossRef]
- Lu, Y.; Abdou, A.M.; Cerhan, J.R.; Morton, L.M.; Severson, R.K.; Davis, S.; Cozen, W.; Rothman, N.; Bernstein, L.; Chanock, S.; et al. Human leukocyte antigen class I and II alleles and overall survival in diffuse large B-cell lymphoma and follicular lymphoma. ScientificWorldJournal 2011, 11, 2062–2070. [Google Scholar] [CrossRef] [PubMed]
- Alcoceba, M.; Sebastian, E.; Marin, L.; Balanzategui, A.; Sarasquete, M.E.; Chillón, M.C.; Jiménez, C.; Puig, N.; Corral, R.; Pardal, E.; et al. HLA specificities are related to development and prognosis of diffuse large B-cell lymphoma. Blood 2013, 122, 1448–1454. [Google Scholar] [CrossRef] [PubMed]
- García-, M.; Alcoceba, M.; López-Parra, M.; Puig, N.; Antón, A.; Balanzategui, A.; Prieto-Conde, I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; et al. HLA specificities are associated with prognosis in IGHV-mutated CLL-like high-count monoclonal B cell lymphocytosis. PLoS ONE 2017, 12, e0172978. [Google Scholar]
- Wrench, D.; Leighton, P.; Skibola, C.F.; Conde, L.; Cazier, J.B.; Matthews, J.; Iqbal, S.; Carlotti, E.; Bodor, C.; Montoto, S.; et al. SNP rs6457327 in the HLA region on chromosome 6p is predictive of the transformation of follicular lymphoma. Blood 2011, 117, 3147–3150. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef]
- Sarkozy, C.; Huet, S.; Carlton, V.E.; Fabiani, B.; Delmer, A.; Jardin, F.; Delfau-Larue, M.H.; Hacini, M.; Ribrag, V.; Guidez, S.; et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget 2017, 8, 8765–8774. [Google Scholar] [CrossRef]
- Alcoceba, M.; García-Álvarez, M.; Chillón, M.C.; Jiménez, C.; Medina, A.; Antón, A.; Blanco, O.; Díaz, L.G.; Tamayo, P.; González-Calle, V.; et al. Liquid biopsy: A non-invasive approach for Hodgkin lymphoma genotyping. Br. J. Haematol. 2021, 195, 542–551. [Google Scholar] [CrossRef]
- Lakhotia, R.; Melani, C.; Dunleavy, K.; Pittaluga, S.; Saba, N.S.; Lindenberg, L.; Mena, E.; Bergvall, E.; Lucas, A.N.; Jacob, A.P.; et al. Circulating Tumor DNA Predicts Therapeutic Outcome in Mantle Cell Lymphoma. Blood Adv. 2022, 6, 2667–2680. [Google Scholar] [CrossRef]
- Delfau-Larue, M.H.; van der Gucht, A.; Dupuis, J.; Jais, J.P.; Nel, I.; Beldi-Ferchiou, A.; Hamdane, S.; Benmaad, I.; Laboure, G.; Verret, B.; et al. Total metabolic tumor volume, circulating tumor cells, cell-free DNA: Distinct prognostic value in follicular lymphoma. Blood Adv. 2018, 2, 807–816. [Google Scholar] [CrossRef]
- Höpken, U.E. Targeting HDAC3 in CREBBP-Mutant Lymphomas Counterstrikes Unopposed Enhancer Deacetylation of B-cell Signaling and Immune Response Genes. Cancer Discov. 2017, 7, 14–16. [Google Scholar] [CrossRef]
- Morschhauser, F.; Tilly, H.; Chaidos, A.; McKay, P.; Phillips, T.; Assouline, S.; Batlevi, C.L.; Campbell, P.; Ribrag, V.; Damaj, G.L.; et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: An open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020, 21, 1433–1442. [Google Scholar] [CrossRef]
- Patel, A.; Oluwole, O.; Savani, B.; Dholaria, B. Taking a BiTE out of the CAR T space race. Br. J. Haematol. 2021, 195, 689–697. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcoceba, M.; García-Álvarez, M.; Okosun, J.; Ferrero, S.; Ladetto, M.; Fitzgibbon, J.; García-Sanz, R. Genetics of Transformed Follicular Lymphoma. Hemato 2022, 3, 615-633. https://doi.org/10.3390/hemato3040042
Alcoceba M, García-Álvarez M, Okosun J, Ferrero S, Ladetto M, Fitzgibbon J, García-Sanz R. Genetics of Transformed Follicular Lymphoma. Hemato. 2022; 3(4):615-633. https://doi.org/10.3390/hemato3040042
Chicago/Turabian StyleAlcoceba, Miguel, María García-Álvarez, Jessica Okosun, Simone Ferrero, Marco Ladetto, Jude Fitzgibbon, and Ramón García-Sanz. 2022. "Genetics of Transformed Follicular Lymphoma" Hemato 3, no. 4: 615-633. https://doi.org/10.3390/hemato3040042
APA StyleAlcoceba, M., García-Álvarez, M., Okosun, J., Ferrero, S., Ladetto, M., Fitzgibbon, J., & García-Sanz, R. (2022). Genetics of Transformed Follicular Lymphoma. Hemato, 3(4), 615-633. https://doi.org/10.3390/hemato3040042









