Transcriptome Sequencing of the Striped Cucumber Beetle, Acalymma vittatum (F.), Reveals Numerous Sex-Specific Transcripts and Xenobiotic Detoxification Genes
Abstract
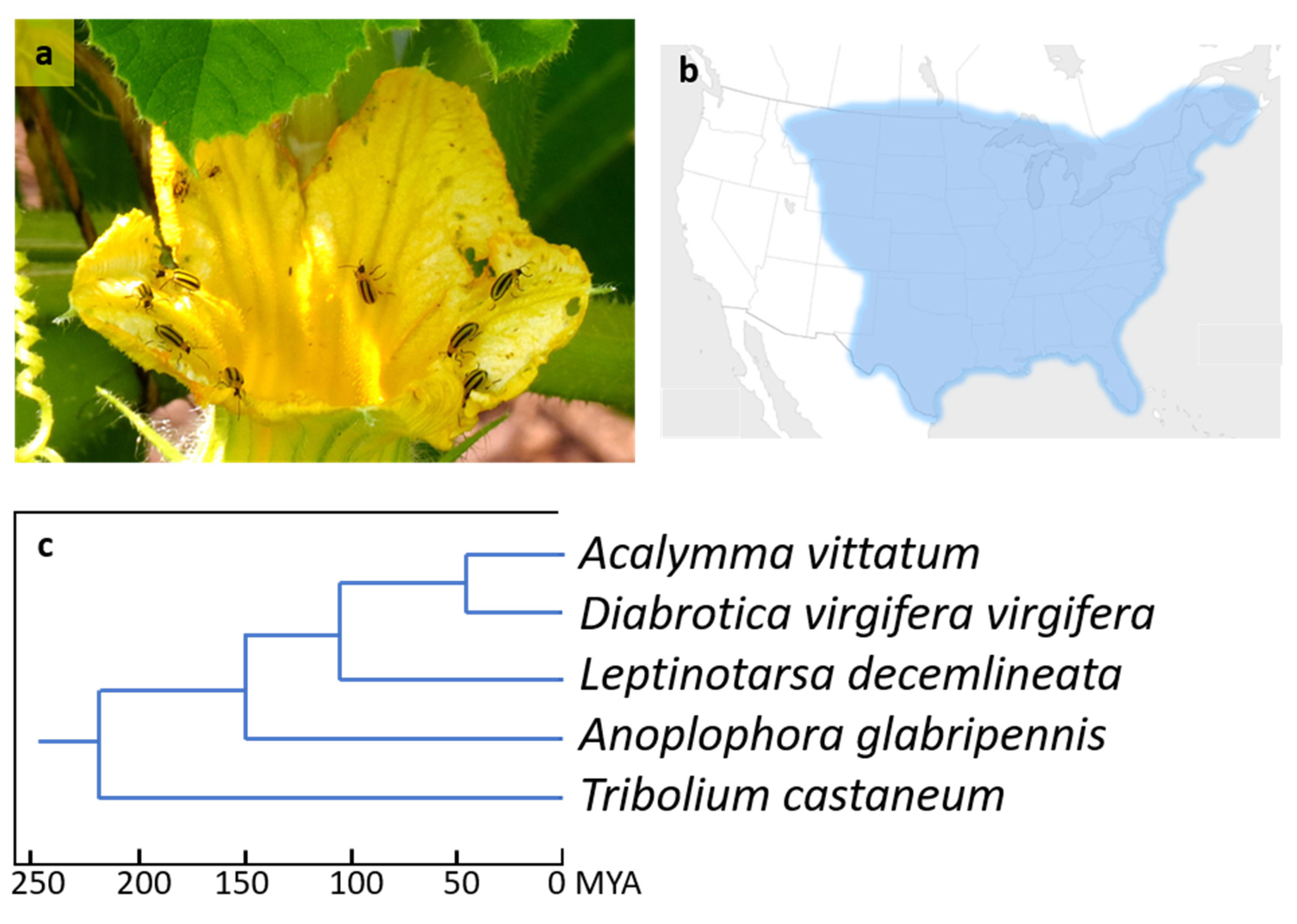
1. Introduction
2. Materials and Methods
3. Results
3.1. Sex-Specific Gene Expression
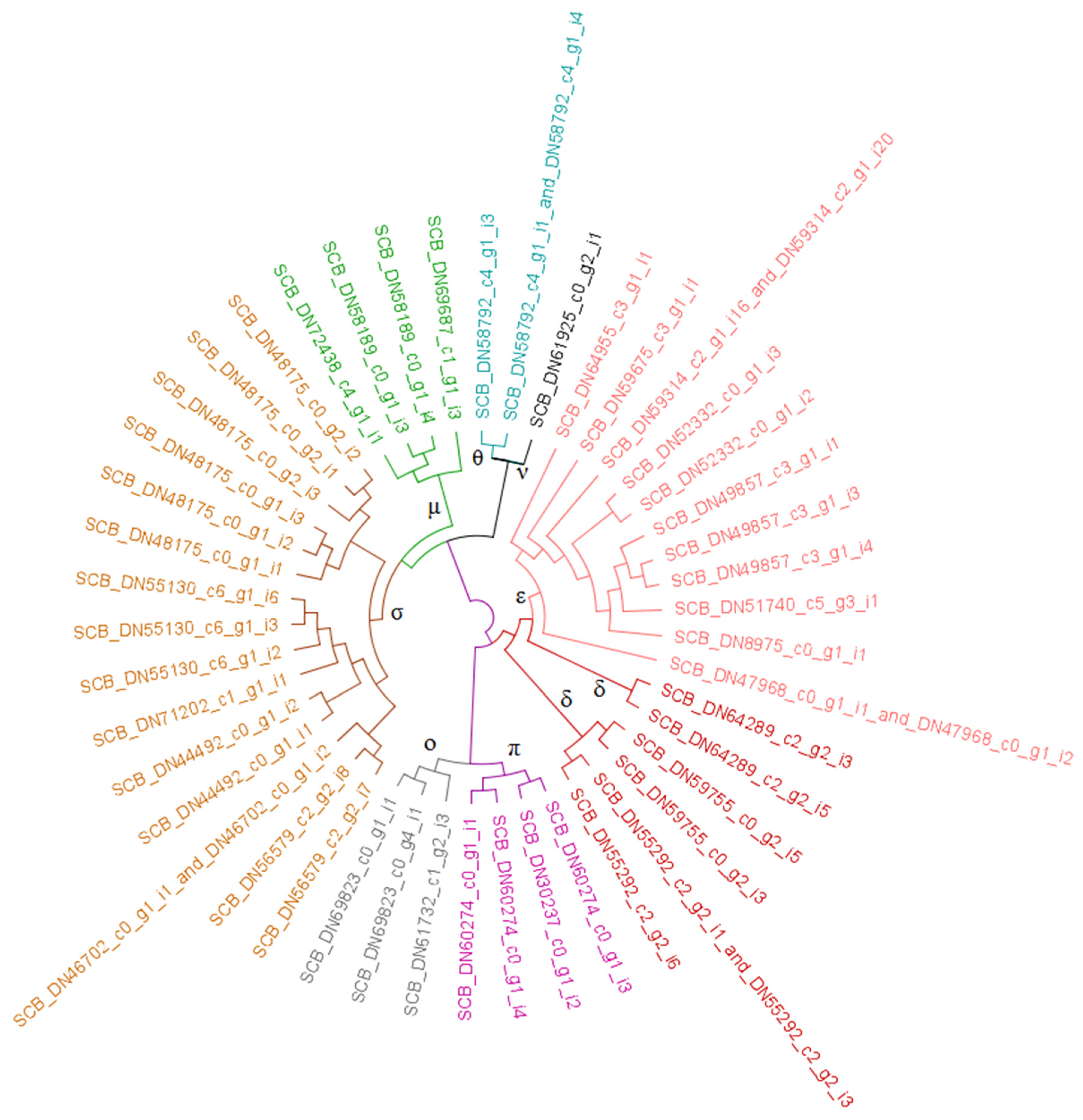
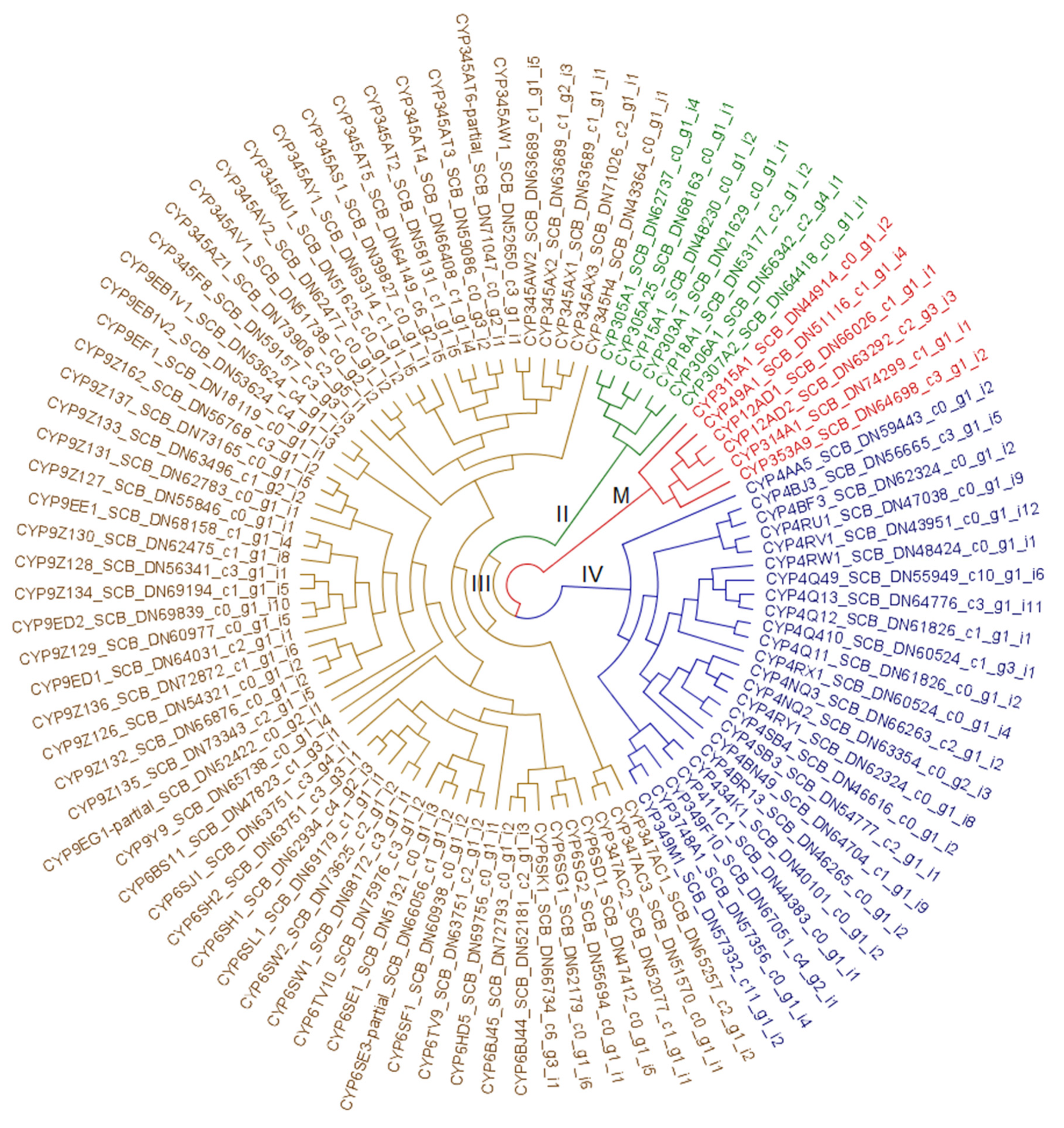
3.2. Resistome Characterization
3.2.1. Glutathione S-Transferases
3.2.2. Carboxylesterases
3.2.3. Cytochrome P450 Monooxygenases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Capinera, J.L. Handbook of Vegetable Pests, 2nd ed.; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Haber, A.I.; Wallingford, A.K.; Grettenberger, I.M.; Ramirez Bonilla, J.P.; Vinchesi-Vahl, A.C.; Weber, D.C. Striped cucumber beetle (Acalymma vittatum (F.)) and Western striped cucumber beetle (Acalymma trivittatum (Mannerheim)) (Coleoptera: Chrysomelidae): Pest Profile. J. Integr. Pest Manag. 2020, in press. [Google Scholar]
- Rojas, E.S.; Batzer, J.C.; Beattie, G.A.; Fleischer, S.J.; Shapiro, L.R.; Williams, M.A.; Bessin, R.; Bruton, B.D.; Boucher, T.J.; Jesse, L.C.H.; et al. Bacterial wilt of cucurbits: Resurrecting a classic pathosystem. Plant Dis. 2015, 99, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Smyth, R.R.; Hoffmann, M.P. A male-produced aggregation pheromone facilitating Acalymma vittatum [F.] (Coleoptera: Chrysomelidae) early-season host plant colonization. J. Insect Behav. 2003, 16, 347–359. [Google Scholar] [CrossRef]
- Morris, B.D.; Smyth, R.R.; Foster, S.P.; Hoffmann, M.P.; Roelofs, W.L.; Franke, S.; Francke, W. Vittatalactone, a beta-lactone from the striped cucumber beetle, Acalymma vittatum. J. Nat. Prod. 2005, 68, 26–30. [Google Scholar] [CrossRef]
- Adler, L.S.; Hazzard, R.V. Comparison of perimeter trap crop varieties: Effects on herbivory, pollination, and yield in butternut squash. Environ. Entomol. 2009, 38, 207–215. [Google Scholar] [CrossRef]
- Cavanagh, A.; Hazzard, R.; Adler, L.S.; Boucher, J. Using trap crops for control of Acalymma vittatum (Coleoptera: Chrysomelidae) reduces insecticide use in butternut squash. J. Econ. Entomol. 2009, 102, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Snyder, W.E. Managing Cucumber Beetles in Organic Farming Systems; Department of Entomology, Washington State University Pullman: Pullman, WA, USA, 2015. [Google Scholar]
- Weber, D.C. Field attraction of striped cucumber beetles to a synthetic vittatalactone mixture. J. Econ. Entomol. 2018, 111, 2988–2991. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Niu, J.; Nji Tizi Taning, C. RNAi in insects: A revolution in fundamental research and pest control applications. Insects 2020, 11, 415. [Google Scholar] [CrossRef]
- Gundersen-Rindal, D.E.; Adrianos, S.L.; Allen, M.L.; Becnel, J.J.; Chen, Y.P.; Choi, M.-Y.; Estep, A.; Evans, J.D.; Garczynski, S.F.; Geib, S.M.; et al. Arthropod genomics research in the United States Department of Agriculture Agricultural Research Service: Applications of RNA interference and CRISPR gene editing technologies in pest control. Trends Entomol. 2017, 13, 109–137. [Google Scholar]
- Sparks, M.E.; Blackburn, M.B.; Kuhar, D.; Gundersen-Rindal, D.E. Transcriptome of the Lymantria dispar (gypsy moth) larval midgut in response to infection by Bacillus thuringiensis. PLoS ONE 2013, 8, e61190. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.E.; Shelby, K.S.; Kuhar, D.; Gundersen-Rindal, D.E. Transcriptome of the invasive brown marmorated stink bug, Halyomorpha halys (Stål) (Heteroptera: Pentatomidae). PLoS ONE 2014, 9, e111646. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.E.; Rhoades, J.H.; Nelson, D.R.; Kuhar, D.; Lancaster, J.; Lehner, B.; Tholl, D.; Weber, D.C.; Gundersen-Rindal, D.E. A transcriptome survey spanning life stages and sexes of the harlequin bug, Murgantia histrionica. Insects 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.E.; Bansal, R.; Benoit, J.B.; Blackburn, M.B.; Chao, H.; Chen, M.; Cheng, S.; Childers, C.; Dinh, H.; Doddapaneni, H.V.; et al. Brown marmorated stink bug, Halyomorpha halys (Stål), genome: Putative underpinnings of polyphagy, insecticide resistance potential and biology of a top worldwide pest. BMC Genomics 2020, 21, 227. [Google Scholar] [CrossRef]
- Allen, M.L.; Rhoades, J.H.; Sparks, M.E.; Grodowitz, M.J. Differential gene expression in red imported fire ant (Solenopsis invicta) (Hymenoptera: Formicidae) larval and pupal stages. Insects 2018, 9, 185. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Bansal, R.; Kerstetter, R.A.; Chen, M.; Carroll, M.; Flannagan, R.; Clark, T.; Goldman, B.S.; Michel, A.P. Western corn rootworm (Diabrotica virgifera virgifera) transcriptome assembly and genomic analysis of population structure. BMC Genomics 2014, 15, 195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-Q.; Che, L.-H.; Li, Y.; Liang, D.; Pang, H.; Ślipiński, A.; Zhang, P. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commun. 2018, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Eben, A.; Espinosa de Los Monteros, A. Tempo and mode of evolutionary radiation in Diabroticina beetles (genera Acalymma, Cerotoma, and Diabrotica). Zookeys 2013, 332, 207–321. [Google Scholar] [CrossRef][Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-Seq: Reference generation and analysis with Trinity. Nat. Protoc. 2013, 8. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 2015, 4, 1521. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Ruotti, V.; Stewart, R.M.; Thomson, J.A.; Dewey, C.N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 2010, 26, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- McKenna, D.D.; Scully, E.D.; Pauchet, Y.; Hoover, K.; Kirsch, R.; Geib, S.M.; Mitchell, R.F.; Waterhouse, R.M.; Ahn, S.-J.; Arsala, D.; et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2016, 17, 227. [Google Scholar] [CrossRef]
- Schoville, S.D.; Chen, Y.H.; Andersson, M.N.; Benoit, J.B.; Bhandari, A.; Bowsher, J.H.; Brevik, K.; Cappelle, K.; Chen, M.-J.M.; Childers, A.K.; et al. A model species for agricultural pest genomics: The genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 2018, 8, 1931. [Google Scholar] [CrossRef]
- Tribolium Genome Sequencing Consortium; Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Beeman, R.W.; Brown, S.J.; et al. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree, Version 1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, UK, 2006; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 31 July 2020).
- Kapli, P.; Lutteropp, S.; Flouri, T. Newick-Tools: A Novel Software for Simulating and Processing Phylogenetic Trees, Version 0.0.1; Heidelberg Institute for Theoretical Studies: Heidelberg, Germany, 2018; Available online: https://github.com/xflouris/newick-tools/ (accessed on 3 September 2020).
- Wernersson, R. Virtual Ribosome—A comprehensive DNA translation tool with support for integration of sequence feature annotation. Nucleic Acids Res 2006, 34, W385–W388. [Google Scholar] [CrossRef]
- Zhu, F.; Moural, T.W.; Nelson, D.R.; Palli, S.R. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple cytochrome P450s. Sci Rep. 2016, 6, 20421. [Google Scholar] [CrossRef]
- Shi, H.; Pei, L.; Gu, S.; Zhu, S.; Wang, Y.; Zhang, Y.; Li, B. Glutathione S-transferase (GST) genes in the red flour beetle, Tribolium castaneum, and comparative analysis with five additional insects. Genomics 2012, 100, 327–335. [Google Scholar] [CrossRef]
- Chelvanayagam, G.; Parker, M.W.; Board, P. Fly fishing for GSTs: A unified nomenclature for mammalian and insect glutathione transferases. Chemico-Biol. Interact. 2001, 133, 256–260. [Google Scholar]
- Friedman, R. Genomic organization of the glutathione S-transferase family in insects. Mol. Phylogenet. Evol. 2011, 61, 924–932. [Google Scholar] [CrossRef]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
- Bartelt, R.J.; Weisleder, D. Polyketide origin of pheromones of Carpophilus davidsoni and C. mutilatus (Coleoptera: Nitidulidae). Bioorg. Med. Chem. 1996, 4, 429–438. [Google Scholar] [CrossRef]
- Pankewitz, F.; Hilker, M. Polyketides in insects: Ecological role of these widespread chemicals and evolutionary aspects of their biogenesis. Biol. Rev. Camb. Philos. Soc. 2008, 83, 209–226. [Google Scholar] [CrossRef]
- Deyrup, S.T.; Eckman, L.E.; Lucadamo, E.E.; McCarthy, P.H.; Knapp, J.C.; Smedley, S.R. Antipredator activity and endogenous biosynthesis of defensive secretion in larval and pupal Delphastus catalinae (Horn) (Coleoptera: Coccinellidae). Chemoecology 2014, 24, 145–157. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef]
- Piel, J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 2002, 99, 14002–14007. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, K.; Engeser, M.; Blunt, J.W.; Munro, M.H.G.; Piel, J. Pederin-type pathways of uncultivated bacterial symbionts: Analysis of O-methyltransferases and generation of a biosynthetic hybrid. J. Am. Chem. Soc. 2009, 131, 2780–2781. [Google Scholar] [CrossRef]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001, 360, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Tu, H.; Shang, Q.; Gao, X.; Liang, P. Molecular cloning and characterization of five glutathione S-transferase genes and promoters from Micromelalopha troglodyta (Graeser) (Lepidoptera: Notodontidae) and their response to tannic acid stress. Insects 2020, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect. Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.; Singh, K.P.; Ali, V.; Behera, S.; Shivam, P.; Das, P.; Dinesh, D.S. Detection and functional characterization of sigma class GST in Phlebotomus argentipes and its role in stress tolerance and DDT resistance. Sci. Rep. 2019, 9, 19636. [Google Scholar] [CrossRef]
- Gawande, N.D.; Subashini, S.; Murugan, M.; Subbarayalu, M. Molecular screening of insecticides with sigma glutathione S-transferases (GST) in cotton aphid Aphis gossypii using docking. Bioinformation 2014, 10, 679–683. [Google Scholar] [CrossRef]
- Hossain, M.D.T.; Yamada, N.; Yamamoto, K. Glutathione-binding site of a Bombyx mori theta-class glutathione transferase. PLoS ONE 2014, 9, e97740. [Google Scholar] [CrossRef] [PubMed]
- Board, P.G. The omega-class glutathione transferases: Structure, function, and genetics. Drug Metab. Rev. 2011, 43, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.J.; Scully, E.D.; Geib, S.M.; Hoover, K. Contrasting diets reveal metabolic plasticity in the tree-killing beetle, Anoplophora glabripennis (Cerambycidae: Lamiinae). Sci. Rep. 2016, 6, 33813. [Google Scholar] [CrossRef]
- Zhang, J.; Li, D.; Ge, P.; Guo, Y.; Zhu, K.Y.; Ma, E.; Zhang, J. Molecular and functional characterization of cDNAs putatively encoding carboxylesterases from the migratory locust, Locusta migratoria. PLoS ONE 2014, 9, e94809. [Google Scholar] [CrossRef] [PubMed]
- Oakeshott, J.G.; Claudianos, C.; Campbell, P.M.; Newcomb, R.D.; Russell, R.J. Biochemical genetics and genomics of insect esterases. In Insect Pharmacology: Channels, Receptors, Toxins and Enzymes; Gilbert, L.I., Gill, S.S., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; pp. 229–306. [Google Scholar]
- Menozzi, P.; Shi, M.A.; Lougarre, A.; Tang, Z.H.; Fournier, D. Mutations of acetylcholinesterase which confer insecticide resistance in Drosophila melanogaster populations. BMC Evol. Biol. 2004, 4, 4. [Google Scholar] [CrossRef]
- Villatte, F.; Ziliani, P.; Marcel, V.; Menozzi, P.; Fournier, D. A high number of mutations in insect acetylcholinesterase may provide insecticide resistance. Pestic. Biochem. Physiol. 2000, 67, 95–102. [Google Scholar] [CrossRef]
- Stankovic, S.; Kostic, M. Role of carboxylesterases (ALiE) regarding resistance to insecticides: Case study of Colorado potato beetle (Leptinotarsa decemlineata Say). In Insect Physiology and Ecology; IntechOpen: London, UK, 2017; pp. 159–178. [Google Scholar]
- Kakugawa, S.; Langton, P.F.; Zebisch, M.; Howell, S.; Chang, T.-H.; Liu, Y.; Feizi, T.; Bineva, G.; O’Reilly, N.; Snijders, A.P.; et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature 2015, 519, 187–192. [Google Scholar] [CrossRef]
- Rewitz, K.F.; O’Connor, M.B.; Gilbert, L.I. Molecular evolution of the insect Halloween family of cytochrome P450s: Phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 2007, 37, 741–753. [Google Scholar] [CrossRef]
- Qiu, Y.; Tittiger, C.; Wicker-Thomas, C.; Le Goff, G.; Young, S.; Wajnberg, E.; Fricaux, T.; Taquet, N.; Blomquist, G.J.; Feyereisen, R. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 14858–14863. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Z.; Zha, S.; Wang, Y.; Jiang, W.; Liao, Y.; Song, Z.; Qi, Z.; Yin, Y. De novo transcriptome and expression profile analysis to reveal genes and pathways potentially involved in cantharidin biosynthesis in the blister beetle Mylabris cichorii. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Luo, C.; Li, Y.; Liao, H.; Yang, Y. De novo transcriptome assembly of the bamboo snout beetle Cyrtotrachelus buqueti reveals ability to degrade lignocellulose of bamboo feedstock. Biotechnol. Biofuels 2018, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Cai, Y.; Zhuo, Z.; Yang, W.; Yang, C.; Zhang, J.; Yang, Y.; Wang, B.; Guan, F. Transcriptome analysis in different developmental stages of Batocera horsfieldi (Coleoptera: Cerambycidae) and comparison of candidate olfactory genes. PLoS ONE 2018, 13, e0192730. [Google Scholar] [CrossRef]
- Khan, S.A.; Eggleston, H.; Myles, K.M.; Adelman, Z.N. Differentially and co-expressed genes in embryo, germ-line and somatic tissues of Tribolium castaneum. G3 (Bethesda) 2019, 9, 2363–2373. [Google Scholar] [CrossRef]
- Immonen, E.; Sayadi, A.; Bayram, H.; Arnqvist, G. Mating changes sexually dimorphic gene expression in the seed beetle Callosobruchus maculatus. Genome Biol. Evol. 2017, 9, 677–699. [Google Scholar] [CrossRef]
- Zotti, M.J.; Smagghe, G. RNAi technology for insect management and protection of beneficial insects from diseases: Lessons, challenges and risk assessments. Neotrop. Entomol. 2015, 44, 197–213. [Google Scholar] [CrossRef] [PubMed]

| Male (♂) | Female (♀) | Pooled and Normalized | |||||
|---|---|---|---|---|---|---|---|
| Biorep A | Biorep B | Biorep C | Biorep A | Biorep B | Biorep C | ||
| read pairs | 87,090,530 | 104,200,730 | 104,805,615 | 97,979,570 | 83,184,389 | 79,066,591 | 32,489,733 |
| R1 bases | 348,362,120 | 416,802,920 | 419,222,460 | 391,918,280 | 332,737,556 | 316,266,364 | 4,887,723,739 |
| R2 bases | 348,362,120 | 416,802,920 | 419,222,460 | 391,918,280 | 332,737,556 | 316,266,364 | 4,896,385,761 |
 ~ male,
~ male,  ~ female). Suggested gene function, as inferred on the basis of hits identified in NCBI NR, is also presented (NR Hits/ Inferred Function).
~ female). Suggested gene function, as inferred on the basis of hits identified in NCBI NR, is also presented (NR Hits/ Inferred Function).
 ~ male,
~ male,  ~ female). Suggested gene function, as inferred on the basis of hits identified in NCBI NR, is also presented (NR Hits/ Inferred Function).
~ female). Suggested gene function, as inferred on the basis of hits identified in NCBI NR, is also presented (NR Hits/ Inferred Function).| TPM | TPM | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Fold ∆ | P-Adj | ♂ A | ♂ B | ♂ C | ♀ A | ♀ B | ♀ C | NR Hits/ Inferred Function |
| DN48906_c127_g1 | 1.39 × 101 | 4.86 × 10−69 | 2342.39 | 3149.43 | 2719.37 | 0.25 | 0.38 | 0.11 | none |
| DN48785_c0_g1 | 1.37 × 101 | 3.94 × 10−66 | 3550.32 | 4564.73 | 4809.53 | 0.40 | 0.55 | 0.43 | none |
| DN52678_c6_g1 | 1.39 × 101 | 3.65 × 10−53 | 1201.27 | 1448.84 | 1526.59 | 0.14 | 0.12 | 0.19 | antichymotrypsin-2; alaserpin |
| DN47640_c7_g1 | 1.36 × 101 | 1.85 × 10−51 | 1503.71 | 1670.18 | 1706.32 | 0.11 | 0.28 | 0.11 | none |
| DN49285_c6_g1 | 1.36 × 101 | 4.15 × 10−49 | 1353.96 | 1585.53 | 1467.49 | 0.07 | 0.05 | 0.19 | uncharacterized protein |
| DN52414_c3_g1 | 1.23 × 101 | 1.23 × 10−47 | 3039.40 | 3952.92 | 4415.55 | 0.74 | 0.86 | 0.63 | none |
| DN69908_c0_g1 | 1.00 × 101 | 9.96 × 10−25 | 9.97 | 7.91 | 5.41 | 0.02 | 0.00 | 0.01 | ELKS/Rab6-interacting/CAST; kinectin |
| DN54481_c1_g3 | 9.47 × 100 | 1.65 × 10−24 | 20.37 | 24.23 | 22.57 | 0.04 | 0.03 | 0.03 | IQ domain-containing protein D |
| DN67865_c1_g1 | 1.19 × 101 | 9.71 × 10−24 | 1174.38 | 1563.92 | 1652.00 | 0.08 | 0.28 | 0.35 | cysteine rich trypsin inhibitor |
| DN70798_c0_g2 | 1.08 × 101 | 3.29 × 10−23 | 223.26 | 242.80 | 304.90 | 0.00 | 0.16 | 0.25 | zinc metalloproteinase |
| DN69207_c33_g1 | −1.12 × 101 | 1.88 × 10−93 | 2.90 | 2.95 | 1.39 | 4108.93 | 10,335.52 | 4487.69 | vitellogenin |
| DN54794_c0_g1 | −1.18 × 101 | 1.82 × 10−52 | 0.07 | 0.27 | 0.14 | 797.99 | 1193.92 | 401.69 | vitellogenin 1 |
| DN66165_c2_g1 | −1.28 × 101 | 5.76 × 10−50 | 0.03 | 0.11 | 0.08 | 735.01 | 809.74 | 454.49 | triacylglycerol lipase |
| DN59442_c0_g1 | −1.42 × 101 | 4.16 × 10−36 | 0.05 | 0.04 | 0.08 | 1755.04 | 1544.04 | 1398.17 | none |
| DN73776_c0_g1 | −8.62 × 100 | 1.39 × 10−34 | 0.07 | 0.06 | 0.05 | 25.42 | 27.33 | 23.90 | vitellogenin receptor |
| DN61697_c0_g1 | −1.58 × 101 | 3.10 × 10−34 | 0.00 | 0.01 | 0.00 | 469.19 | 414.59 | 238.52 | lysosomal aspartic protease; cathepsin D |
| DN50160_c11_g2 | −2.02 × 101 | 1.87 × 10−33 | 0.00 | 0.00 | 0.00 | 2541.30 | 4509.76 | 5446.56 | glycoside hydrolase family 1 |
| DN73353_c1_g2 | −8.37 × 100 | 1.32 × 10−32 | 2.28 | 1.25 | 1.80 | 191.16 | 167.25 | 134.58 | sphingomyelin phosphodiesterase |
| DN47397_c1_g1 | −1.90 × 101 | 8.41 × 10−32 | 0.00 | 0.00 | 0.00 | 1315.10 | 2893.93 | 391.33 | none |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sparks, M.E.; Nelson, D.R.; Haber, A.I.; Weber, D.C.; Harrison, R.L. Transcriptome Sequencing of the Striped Cucumber Beetle, Acalymma vittatum (F.), Reveals Numerous Sex-Specific Transcripts and Xenobiotic Detoxification Genes. BioTech 2020, 9, 21. https://doi.org/10.3390/biotech9040021
Sparks ME, Nelson DR, Haber AI, Weber DC, Harrison RL. Transcriptome Sequencing of the Striped Cucumber Beetle, Acalymma vittatum (F.), Reveals Numerous Sex-Specific Transcripts and Xenobiotic Detoxification Genes. BioTech. 2020; 9(4):21. https://doi.org/10.3390/biotech9040021
Chicago/Turabian StyleSparks, Michael E., David R. Nelson, Ariela I. Haber, Donald C. Weber, and Robert L. Harrison. 2020. "Transcriptome Sequencing of the Striped Cucumber Beetle, Acalymma vittatum (F.), Reveals Numerous Sex-Specific Transcripts and Xenobiotic Detoxification Genes" BioTech 9, no. 4: 21. https://doi.org/10.3390/biotech9040021
APA StyleSparks, M. E., Nelson, D. R., Haber, A. I., Weber, D. C., & Harrison, R. L. (2020). Transcriptome Sequencing of the Striped Cucumber Beetle, Acalymma vittatum (F.), Reveals Numerous Sex-Specific Transcripts and Xenobiotic Detoxification Genes. BioTech, 9(4), 21. https://doi.org/10.3390/biotech9040021







