RNA Polymerase II Activity and Nuclear Actin: Possible Roles of Nuclear Tropomyosin, Troponin and Ca2+ in Transcription in Striated Muscle Myocyte Nuclei
Abstract
1. Eukaryotic Transcription and Its Regulation
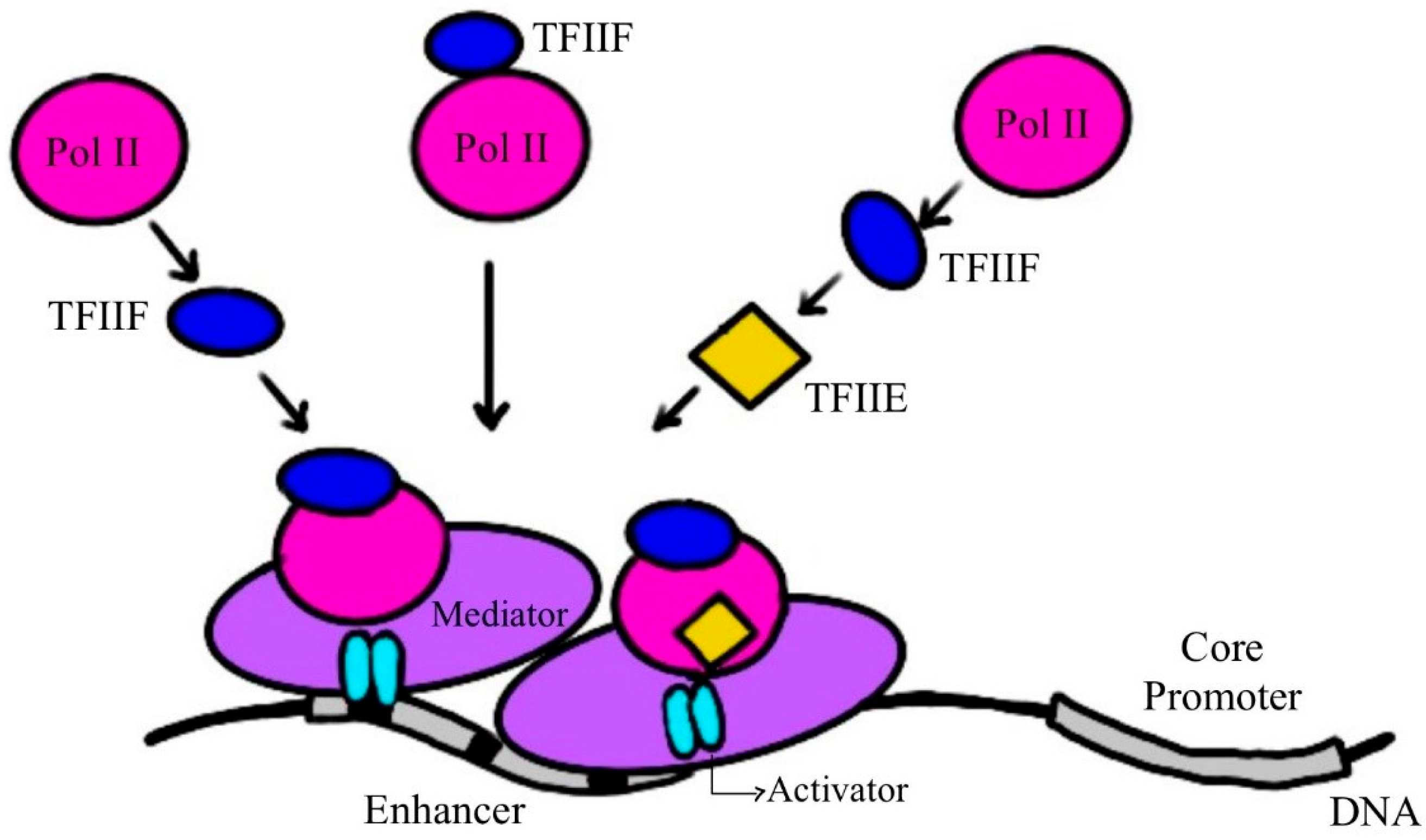
2. RNA Polymerase, the Molecular Machine Responsible for Transcription
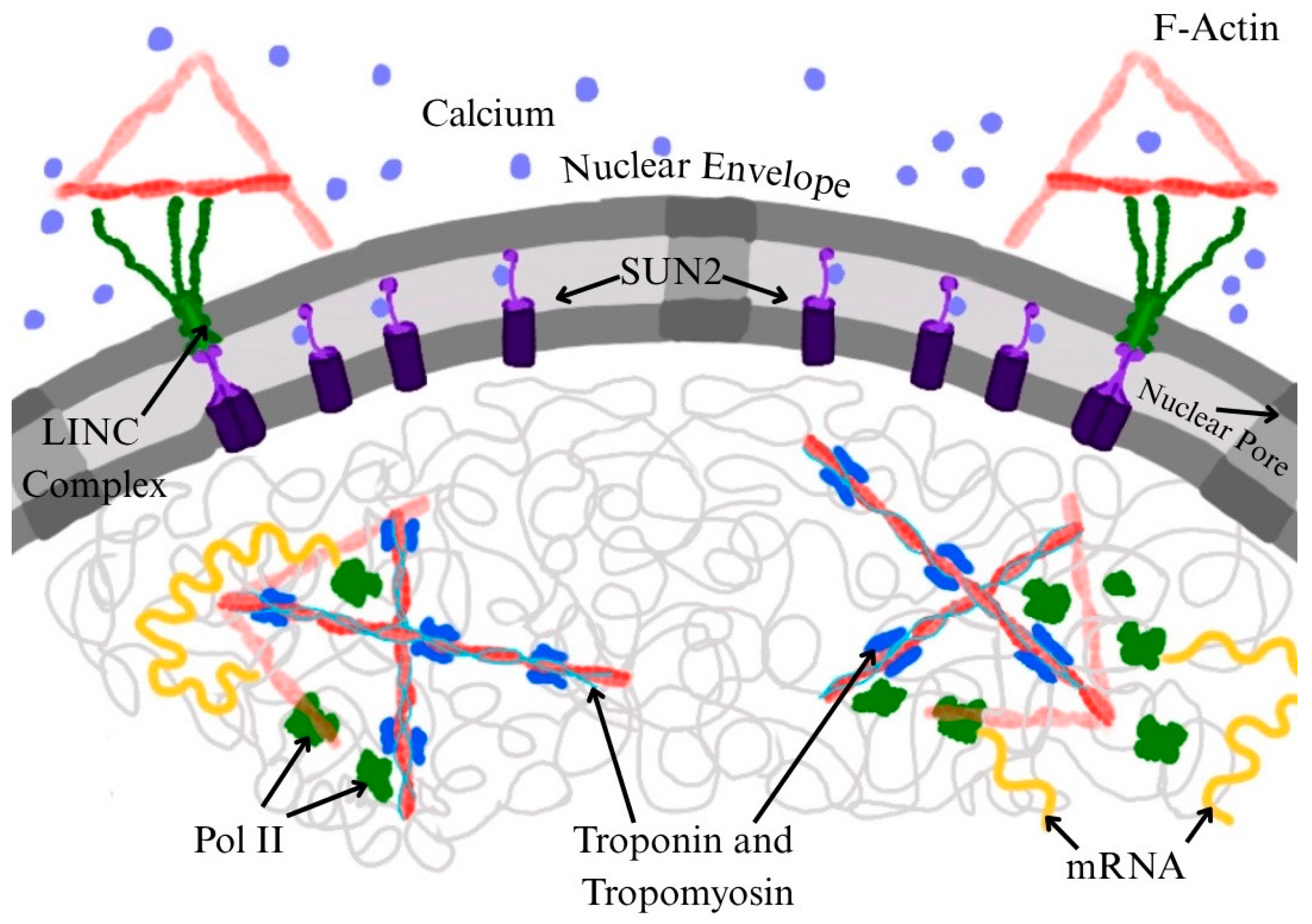
3. Requirement of Nuclear Actin for RNA Pol II Function
4. Troponin and Tropomyosin as Potential Modulators of Nuclear Actin and Transcription
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Arp2/3 | Actin-related proteins 2/3 |
| ARP4 | Actin-related protein 4 |
| BAF complex | BRG/BRM-associated factor complex |
| CaMKII | Ca2+-calmodulin-dependent protein kinase II |
| CaN | Calcineurin |
| ChromExM | Chromatin expansion microscopy |
| CREB | Cyclic-AMP response element-binding |
| CTD | Disordered carboxy-terminal domain of RNA Pol II |
| DNA | Deoxyribonucleic acid |
| ERK1/2 | Extracellular signal-related kinases 1 and 2 |
| F-actin | Filamentous (polymerized) actin |
| G-actin | Globular (monomeric) actin |
| GTP | Guanosine triphosphate |
| HDAC | Histone deacetylase |
| INM | Inner nuclear membrane |
| IP3 | Inositol trisphosphate |
| LINC | Linker of nucleoskeleton and cytoskeleton |
| MAL | Megakaryocytic acute leukemia protein |
| MAPK | Mitogen-activated protein kinase |
| miRNAs | microRNAs |
| MRFs | Myogenic regulatory factors |
| mRNA | Messenger ribonucleic acid |
| MyoD | Myoblast determination protein |
| NFAT | Nuclear factor of activated T-cells |
| NM1 | Nuclear myosin 1 |
| NMVI | Nuclear myosin VI |
| PIC | Pre-initiation complex for transcription |
| Pol I | RNA polymerase I |
| Pol II | RNA polymerase II |
| Pol III | RNA polymerase III |
| Pol IV | RNA polymerase IV |
| Pol V | RNA polymerase V |
| pre-mRNA | Pre-messenger RNA |
| P-TEFb | Positive transcription elongation factor b |
| rDNA | Gene for a ribosomal RNA |
| RNA | Ribonucleic acid |
| rRNA | Ribosomal ribonucleic acid |
| snRNAs | small nuclear RNAs |
| SRF | Serum response factor |
| SUMO | Small ubiquitin-like modifier protein |
| SWI/SNF | switch/sucrose non-fermentable chromatin-remodeling complex |
| TBP | TATA-binding protein |
| TFs | Transcription factors |
| Tn | Troponin |
| TnC | Troponin C subunit of troponin |
| TnI | Troponin I subunit of troponin |
| TnT | Troponin T subunit of troponin |
| Tm | Tropomyosin |
| tRNA | Transfer ribonucleic acid |
| Ubc9 | ubiquitin conjugating enzyme Ubc9 (E2I) |
| YAP | Yes-associated protein |
References
- Piccolino, M. Biological machines: From mills to molecules. Nat. Rev. Mol. Cell Biol. 2000, 1, 149–153. [Google Scholar] [CrossRef] [PubMed]
- van den Heuvel, M.G.L.; Dekker, C. Motor proteins at work for nanotechnology. Science 2007, 317, 333–336. [Google Scholar] [CrossRef]
- Cramer, P. A Tale of Chromatin and Transcription in 100 Structures. Cell 2014, 159, 985–994. [Google Scholar] [CrossRef]
- Trivedi, D.V.; Nag, S.; Spudich, A.; Ruppel, K.M.; Spudich, J.A. The Myosin Family of Mechanoenzymes: From Mechanisms to Therapeutic Approaches. Annu. Rev. Biochem. 2020, 89, 667–693. [Google Scholar] [CrossRef]
- Roeder, R.G. 50+ years of eukaryotic transcription: An expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 2019, 26, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Pownall, M.E.; Miao, L.; Vejnar, C.E.; M’Saad, O.; Sherrard, A.; Frederick, M.A.; Benitez, M.D.J.; Boswell, C.W.; Zaret, K.S.; Bewersdorf, J.; et al. Chromatin expansion microscopy reveals nanoscale organization of transcription and chromatin. Science 2023, 381, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Stasevich, T.J.; Kimura, H. An expanded view of transcription. Science 2023, 381, 26–27. [Google Scholar] [CrossRef]
- Nguyen, V.Q.; Ranjan, A.; Liu, S.; Tang, X.; Ling, Y.H.; Wisniewski, J.; Mizuguchi, G.; Li, K.Y.; Jou, V.; Zheng, Q.; et al. Spatiotemporal coordination of transcription preinitiation complex assembly in live cells. Mol. Cell 2021, 81, 3560–3575.e6. [Google Scholar] [CrossRef]
- Baek, I.; Friedman, L.J.; Gelles, J.; Buratowski, S. Single-molecule studies reveal branched pathways for activator-dependent assembly of RNA polymerase II pre-initiation complexes. Mol. Cell 2021, 81, 3576–3588.e6. [Google Scholar] [CrossRef]
- Ling, Y.H.; Ye, Z.; Liang, C.; Yu, C.; Park, G.; Corden, J.L.; Wu, C. Disordered C-terminal domain drives spatiotemporal confinement of RNAPII to enhance search for chromatin targets. Nat. Cell Biol. 2024, 26, 581–592. [Google Scholar] [CrossRef]
- Casamassimi, A.; Ciccodicola, A. Transcriptional Regulation: Molecules, Involved Mechanisms, and Misregulation. Int. J. Mol. Sci. 2019, 20, 1281. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Song, C.; Tang, H. Transcriptional regulation in cardiovascular diseases. Front. Cardiovasc. Med. 2024, 11, 1360765. [Google Scholar] [CrossRef]
- Fuda, N.J.; Ardehali, M.B.; Lis, J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 2009, 461, 186–192. [Google Scholar] [CrossRef]
- Filipovski, M.; Soffers, J.H.M.; Vos, S.M.; Farnung, L. Structural basis of nucleosome retention during transcription elongation. Science 2022, 376, 1313–1316. [Google Scholar] [CrossRef]
- Olson, E.N. Regulation of muscle transcription by the MyoD family. The heart of the matter. Circ. Res. 1993, 72, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.N. Signal transduction pathways that regulate skeletal muscle gene expression. Mol. Endocrinol. 1993, 7, 1369–1378. [Google Scholar] [CrossRef]
- Li, L.; Olson, E.N. Regulation of muscle cell growth and differentiation by the MyoD family of helix-loop-helix proteins. Adv. Cancer Res. 1992, 58, 95–119. [Google Scholar] [CrossRef]
- Braun, T.; Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011, 12, 349–361. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, D.-Z.; Hockemeyer, D.; McAnally, J.; Nordheim, A.; Olson, E.N. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 2004, 428, 185–189. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, D.Z.; Pipes, G.C.; Olson, E.N. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. USA 2003, 100, 7129–7134. [Google Scholar] [CrossRef] [PubMed]
- Estrella, N.L.; Naya, F.J. Transcriptional networks regulating the costamere, sarcomere, and other cytoskeletal structures in striated muscle. Cell. Mol. Life Sci. 2014, 71, 1641–1656. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Han, M.; Bernier, M.; Wen, J.-k. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009, 276, 2669–2685. [Google Scholar] [CrossRef]
- Roeder, R.G.; Rutter, W.J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 1969, 224, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Wu, X.-X.; Fang, C.-L.; Xu, Z.-G.; Zhang, H.-W.; Gao, J.; Zhou, C.-M.; You, L.-L.; Gu, Z.-X.; Mu, W.-H.; et al. Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA. Science 2021, 374, 1579–1586. [Google Scholar] [CrossRef] [PubMed]
- Engel, C.; Sainsbury, S.; Cheung, A.C.; Kostrewa, D.; Cramer, P. RNA polymerase I structure and transcription regulation. Nature 2013, 502, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Obrdlik, A.; Louvet, E.; Kukalev, A.; Naschekin, D.; Kiseleva, E.; Fahrenkrog, B.; Percipalle, P. Nuclear myosin 1 is in complex with mature rRNA transcripts and associates with the nuclear pore basket. FASEB J. 2010, 24, 146–157. [Google Scholar] [CrossRef]
- Turowski, T.W.; Tollervey, D. Transcription by RNA polymerase III: Insights into mechanism and regulation. Biochem. Soc. Trans. 2016, 44, 1367–1375. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Rinn, J.; Guttman, M. RNA Function. RNA and dynamic nuclear organization. Science 2014, 345, 1240–1241. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Rissland, O.S.; Norbury, C.J. The Cid1 poly(U) polymerase. Biochim. Biophys. Acta 2008, 1779, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Schier, A.C.; Taatjes, D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Vorländer, M.K.; Müller, C.W. RNA polymerase I and III: Similar yet unique. Curr. Opin. Struct. Biol. 2017, 47, 88–94. [Google Scholar] [CrossRef]
- Kostrewa, D.; Zeller, M.E.; Armache, K.-J.; Seizl, M.; Leike, K.; Thomm, M.; Cramer, P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 2009, 462, 323–330. [Google Scholar] [CrossRef]
- He, Y.; Yan, C.; Fang, J.; Inouye, C.; Tjian, R.; Ivanov, I.; Nogales, E. Near-atomic resolution visualization of human transcription promoter opening. Nature 2016, 533, 359–365. [Google Scholar] [CrossRef]
- Vos, S.M.; Farnung, L.; Urlaub, H.; Cramer, P. Structure of paused transcription complex Pol II-DSIF-NELF. Nature 2018, 560, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Vos, S.M.; Farnung, L.; Boehning, M.; Wigge, C.; Linden, A.; Urlaub, H.; Cramer, P. Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 2018, 560, 607–612. [Google Scholar] [CrossRef]
- Vos, S.M.; Farnung, L.; Linden, A.; Urlaub, H.; Cramer, P. Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation. Nat. Struct. Mol. Biol. 2020, 27, 668–677. [Google Scholar] [CrossRef]
- Kettenberger, H.; Armache, K.-J.; Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 2004, 16, 955–965. [Google Scholar] [CrossRef]
- Aibara, S.; Schilbach, S.; Cramer, P. Structures of mammalian RNA polymerase II pre-initiation complexes. Nature 2021, 594, 124–128. [Google Scholar] [CrossRef]
- Schier, A.C.; Taatjes, D.J. Everything at once: Cryo-EM yields remarkable insights into human RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2021, 28, 540–543. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Liu, W.; Ren, Y.; Qu, X.; Li, J.; Yin, X.; Xu, Y. Structures of +1 nucleosome-bound PIC-Mediator complex. Science 2022, 378, 62–68. [Google Scholar] [CrossRef]
- Chen, X.; Yin, X.; Li, J.; Wu, Z.; Qi, Y.; Wang, X.; Liu, W.; Xu, Y. Structures of the human Mediator and Mediator-bound preinitiation complex. Science 2021, 372, eabg0635. [Google Scholar] [CrossRef]
- Hou, T.Y.; Kraus, W.L. Come one, come all? Re-evaluating RNA polymerase II pre-initiation complex assembly using single-molecule microscopy. Mol. Cell 2021, 81, 3443–3445. [Google Scholar] [CrossRef]
- Chen, X.; Qi, Y.; Wu, Z.; Wang, X.; Li, J.; Zhao, D.; Hou, H.; Li, Y.; Yu, Z.; Liu, W.; et al. Structural insights into preinitiation complex assembly on core promoters. Science 2021, 372, eaba8490. [Google Scholar] [CrossRef]
- Liu, X.; Bushnell, D.A.; Kornberg, R.D. RNA polymerase II transcription: Structure and mechanism. Biochim. Biophys. Acta 2013, 1829, 2–8. [Google Scholar] [CrossRef]
- Donczew, R.; Hahn, S. Mechanistic Differences in Transcription Initiation at TATA-Less and TATA-Containing Promoters. Mol. Cell Biol. 2018, 38, e00448-17. [Google Scholar] [CrossRef]
- Yella, V.R.; Bansal, M. DNA structural features of eukaryotic TATA-containing and TATA-less promoters. FEBS Open Bio. 2017, 7, 324–334. [Google Scholar] [CrossRef]
- Pugh, B.F.; Tjian, R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991, 5, 1935–1945. [Google Scholar] [CrossRef]
- Harlen, K.M.; Churchman, L.S. The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Çolak, R.; et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef]
- Ruiz-Opazo, N.; Nadal-Ginard, B. α-tropomyosin gene organization: Alternative splicing of duplicated isotype-specific exons accounts for the production of smooth and striated muscle isoforms. J. Biol. Chem. 1987, 262, 4755–4765. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol. Rev. 1996, 76, 371–423. [Google Scholar] [CrossRef]
- Bernstein, S.I.; Milligan, R.A. Fine tuning a molecular motor: The location of alternative domains in the Drosophila myosin head. J. Mol. Biol. 1997, 271, 1–6. [Google Scholar] [CrossRef]
- Perry, S.V. Troponin T: Genetics, properties and function. J. Muscle Res. Cell Motil. 1998, 19, 575–602. [Google Scholar] [CrossRef]
- Perry, S.V. Vertebrate tropomyosin: Distribution, properties and function. J. Muscle Res. Cell Motil. 2001, 22, 5–49. [Google Scholar] [CrossRef]
- Swank, D.M.; Kronert, W.A.; Bernstein, S.I.; Maughan, D.W. Alternative N-terminal regions of Drosophila myosin heavy chain tune muscle kinetics for optimal power output. Biophys. J. 2004, 87, 1805–1814. [Google Scholar] [CrossRef]
- Labeit, S.; Lahmers, S.; Burkart, C.; Fong, C.; McNabb, M.; Witt, S.; Witt, C.; Labeit, D.; Granzier, H. Expression of distinct classes of titin isoforms in striated and smooth muscles by alternative splicing, and their conserved interaction with filamins. J. Mol. Biol. 2006, 362, 664–681. [Google Scholar] [CrossRef]
- Sheng, J.J.; Jin, J.-P. Gene regulation, alternative splicing, and posttranslational modification of troponin subunits in cardiac development and adaptation: A focused review. Front. Physiol. 2014, 5, 165. [Google Scholar] [CrossRef]
- Viita, T.; Kyheröinen, S.; Prajapati, B.; Virtanen, J.; Frilander, M.J.; Varjosalo, M.; Vartiainen, M.K. Nuclear actin interactome analysis links actin to KAT14 histone acetyl transferase and mRNA splicing. J. Cell Sci. 2019, 132, jcs226852. [Google Scholar] [CrossRef]
- Huxley, H.E. The mechanism of muscular contraction. Science 1969, 164, 1356–1366. [Google Scholar] [CrossRef]
- Cooke, R. The mechanism of muscle contraction. CRC Crit. Rev. Biochem. 1986, 21, 53–118. [Google Scholar] [CrossRef]
- Holmes, K.C.; Kabsch, W. Muscle proteins: Actin. Curr. Opin. Struct. Biol. 1991, 1, 270–280. [Google Scholar] [CrossRef]
- Pollard, T.D.; Cooper, J.A. Actin, a central player in cell shape and movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef]
- Risi, C.M.; Pepper, I.; Belknap, B.; Landim-Vieira, M.; White, H.D.; Dryden, K.; Pinto, J.R.; Chase, P.B.; Galkin, V.E. The Structure of the Native Cardiac Thin Filament at Systolic Ca2+ Levels. Proc. Natl. Acad. Sci. USA 2021, 118, e2024288118. [Google Scholar] [CrossRef]
- Risi, C.M.; Belknap, B.; Atherton, J.; Coscarella, I.L.; White, H.D.; Chase, P.B.; Pinto, J.R.; Galkin, V.E. Troponin structural dynamics in the native cardiac thin filament revealed by cryo electron microscopy. J. Mol. Biol. 2024, 436, 168498. [Google Scholar] [CrossRef]
- Ohnishi, T.; Kawamura, H.; Tanaka, Y. Actin and Myosin-like Proteins in the Calf Thymus Cell Nucleus. J. Biochem. 1964, 56, 6–15. [Google Scholar] [CrossRef]
- Ohnishi, T.; Kawamura, H.; Yamamoto, T. Extraction of a Protein Resembling Actin from the Cell Nucleus of the Calf Thymus. J. Biochem. 1963, 54, 298–300. [Google Scholar] [CrossRef]
- Jockusch, B.M.; Schoenenberger, C.-A.; Stetefeld, J.; Aebi, U. Tracking down the different forms of nuclear actin. Trends Cell Biol. 2006, 16, 391–396. [Google Scholar] [CrossRef]
- Pederson, T. As functional nuclear actin comes into view, is it globular, filamentous, or both? J. Cell Biol. 2008, 180, 1061–1064. [Google Scholar] [CrossRef]
- Baarlink, C.; Wang, H.; Grosse, R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 2013, 340, 864–867. [Google Scholar] [CrossRef]
- Feric, M.; Brangwynne, C.P. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 2013, 15, 1253–1259. [Google Scholar] [CrossRef]
- Kelpsch, D.J.; Tootle, T.L. Nuclear Actin: From Discovery to Function. Anat. Rec. 2018, 301, 1999–2013. [Google Scholar] [CrossRef]
- Ulferts, S.; Lopes, M.; Miyamoto, K.; Grosse, R. Nuclear actin dynamics and functions at a glance. J. Cell Sci. 2024, 137, jcs261630. [Google Scholar] [CrossRef]
- Hofmann, W.A.; de Lanerolle, P. Nuclear actin: To polymerize or not to polymerize. J. Cell Biol. 2006, 172, 495–496. [Google Scholar] [CrossRef]
- Kloc, M.; Chanana, P.; Vaughn, N.; Uosef, A.; Kubiak, J.Z.; Ghobrial, R.M. New Insights into Cellular Functions of Nuclear Actin. Biology 2021, 10, 304. [Google Scholar] [CrossRef]
- Serebryannyy, L.A.; Parilla, M.; Annibale, P.; Cruz, C.M.; Laster, K.; Gratton, E.; Kudryashov, D.; Kosak, S.T.; Gottardi, C.J.; de Lanerolle, P. Persistent nuclear actin filaments inhibit transcription by RNA polymerase II. J. Cell Sci. 2016, 129, 3412–3425. [Google Scholar] [CrossRef]
- Hofmann, W.A.; Arduini, A.; Nicol, S.M.; Camacho, C.J.; Lessard, J.L.; Fuller-Pace, F.V.; de Lanerolle, P. SUMOylation of nuclear actin. J. Cell Biol. 2009, 186, 193–200. [Google Scholar] [CrossRef]
- Pollard, T.D. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef]
- Falahzadeh, K.; Banaei-Esfahani, A.; Shahhoseini, M. The potential roles of actin in the nucleus. Cell J. 2015, 17, 7–14. [Google Scholar] [CrossRef]
- Vartiainen, M.K.; Guettler, S.; Larijani, B.; Treisman, R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007, 316, 1749–1752. [Google Scholar] [CrossRef]
- Sen, B.; Xie, Z.; Thomas, M.D.; Pattenden, S.G.; Howard, S.; McGrath, C.; Styner, M.; Uzer, G.; Furey, T.S.; Rubin, J. Nuclear actin structure regulates chromatin accessibility. Nat. Commun. 2024, 15, 4095. [Google Scholar] [CrossRef]
- Vandekerckhove, J.; Weber, K. At least six different actins are expressed in a higher mammal: An analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 1978, 126, 783–802. [Google Scholar] [CrossRef]
- Rajakylä, E.K.; Vartiainen, M.K. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases 2014, 5, e27539. [Google Scholar] [CrossRef]
- Liu, B.; Shuai, K. Regulation of the sumoylation system in gene expression. Curr. Opin. Cell Biol. 2008, 20, 288–293. [Google Scholar] [CrossRef]
- Baek, S.H. A novel link between SUMO modification and cancer metastasis. Cell Cycle 2006, 5, 1492–1495. [Google Scholar] [CrossRef]
- Batut, P.J.; Bing, X.Y.; Sisco, Z.; Raimundo, J.; Levo, M.; Levine, M.S. Genome organization controls transcriptional dynamics during development. Science 2022, 375, 566–570. [Google Scholar] [CrossRef]
- Miralles, F.; Posern, G.; Zaromytidou, A.-I.; Treisman, R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 2003, 113, 329–342. [Google Scholar] [CrossRef]
- Arsenian, S.; Weinhold, B.; Oelgeschläger, M.; Rüther, U.; Nordheim, A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998, 17, 6289–6299. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, M.; Fang, H.; El-Mounayri, O.; Rodenberg, J.M.; Imbalzano, A.N.; Herring, B.P. The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 921–928. [Google Scholar] [CrossRef]
- Miyamoto, K.; Gurdon, J.B. Transcriptional regulation and nuclear reprogramming: Roles of nuclear actin and actin-binding proteins. Cell. Mol. Life Sci. 2013, 70, 3289–3302. [Google Scholar] [CrossRef]
- Alfert, A.; Moreno, N.; Kerl, K. The BAF complex in development and disease. Epigenetics Chromatin 2019, 12, 19. [Google Scholar] [CrossRef]
- Petrusová, J.; Manning, J.; Filipp, D. Envisioning a role for nuclear actin in prophase I spermatocytes. Front. Cell Dev. Biol. 2023, 11, 1295452. [Google Scholar] [CrossRef]
- Wei, M.; Fan, X.; Ding, M.; Li, R.; Shao, S.; Hou, Y.; Meng, S.; Tang, F.; Li, C.; Sun, Y. Nuclear actin regulates inducible transcription by enhancing RNA polymerase II clustering. Sci. Adv. 2020, 6, eaay6515. [Google Scholar] [CrossRef]
- Miyamoto, K.; Gurdon, J.B. Nuclear actin and transcriptional activation. Commun. Integr. Biol. 2011, 4, 582–583. [Google Scholar] [CrossRef]
- Core, L.; Adelman, K. Promoter-proximal pausing of RNA polymerase II: A nexus of gene regulation. Genes Dev. 2019, 33, 960–982. [Google Scholar] [CrossRef]
- Kyheröinen, S.; Prajapati, B.; Sokolova, M.; Schmitz, M.; Viita, T.; Geyer, M.; Vartiainen, M.K. Actin associates with actively elongating genes and binds directly to the Cdk9 subunit of P-TEFb. J. Biol. Chem. 2024, 300, 105698. [Google Scholar] [CrossRef]
- Belin, B.J.; Cimini, B.A.; Blackburn, E.H.; Mullins, R.D. Visualization of actin filaments and monomers in somatic cell nuclei. Mol. Biol. Cell 2013, 24, 982–994. [Google Scholar] [CrossRef]
- Wineland, D.M.; Kelpsch, D.J.; Tootle, T.L. Multiple Pools of Nuclear Actin. Anat. Rec. 2018, 301, 2014–2036. [Google Scholar] [CrossRef]
- Kapoor, P.; Shen, X. Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol. 2014, 24, 238–246. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, S.; Park, J.-I. Nuclear Actin Dynamics in Gene Expression, DNA Repair, and Cancer. In Nuclear, Chromosomal, and Genomic Architecture in Biology and Medicine. Results and Problems in Cell Differentiation; Springer: Cham, Switzerland, 2022; Volume 70, pp. 625–663. [Google Scholar] [CrossRef]
- Saidova, A.A.; Vorobjev, I.A. What Actin and Myosin Do in the Nucleus: New Functions of the Well-Known Proteins. Mol. Biol. 2024, 58, 349–362. [Google Scholar] [CrossRef]
- de Lanerolle, P.; Serebryannyy, L. Nuclear actin and myosins: Life without filaments. Nat. Cell Biol. 2011, 13, 1282–1288. [Google Scholar] [CrossRef]
- de Lanerolle, P. Nuclear actin and myosins at a glance. J. Cell Sci. 2012, 125 Pt 21, 4945–4949. [Google Scholar] [CrossRef]
- Philimonenko, V.V.; Zhao, J.; Iben, S.; Dingova, H.; Kysela, K.; Kahle, M.; Zentgraf, H.; Hofmann, W.A.; de Lanerolle, P.; Hozak, P.; et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 2004, 6, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Hu, Y.; Belmont, A.S. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. 2014, 24, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef]
- Pestic-Dragovich, L.; Stojiljkovic, L.; Philimonenko, A.A.; Nowak, G.; Ke, Y.; Settlage, R.E.; Shabanowitz, J.; Hunt, D.F.; Hozak, P.; de Lanerolle, P. A myosin I isoform in the nucleus. Science 2000, 290, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Vreugde, S.; Ferrai, C.; Miluzio, A.; Hauben, E.; Marchisio, P.C.; Crippa, M.P.; Bussi, M.; Biffo, S. Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Mol. Cell 2006, 23, 749–755. [Google Scholar] [CrossRef]
- Hari-Gupta, Y.; Fili, N.; Dos Santos, Á.; Cook, A.W.; Gough, R.E.; Reed, H.C.W.; Wang, L.; Aaron, J.; Venit, T.; Wait, E.; et al. Myosin VI regulates the spatial organisation of mammalian transcription initiation. Nat. Commun. 2022, 13, 1346. [Google Scholar] [CrossRef]
- Asumda, F.Z.; Chase, P.B. Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation 2012, 83, 106–115. [Google Scholar] [CrossRef]
- Chase, P.B.; Szczypinski, M.P.; Soto, E.P. Nuclear tropomyosin and troponin in striated muscle: New roles in a new locale? J. Muscle Res. Cell Motil. 2013, 34, 275–284. [Google Scholar] [CrossRef]
- Arifulin, E.A.; Sheval, E.V. Non-Canonical Localization of Cardiac Troponins: Expanding Functions or Causing Pathologies? Int. J. Mol. Sci. 2024, 25, 3117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Birbrair, A.; Wang, Z.M.; Taylor, J.; Messi, M.L.; Delbono, O. Troponin T nuclear localization and its role in aging skeletal muscle. Age 2013, 35, 353–370. [Google Scholar] [CrossRef]
- Johnston, J.R.; Chase, P.B.; Pinto, J.R. Troponin through the looking-glass: Emerging roles beyond regulation of striated muscle contraction. Oncotarget 2018, 9, 1461–1482. [Google Scholar] [CrossRef]
- Gordon, A.M.; Homsher, E.; Regnier, M. Regulation of contraction in striated muscle. Physiol. Rev. 2000, 80, 853–924. [Google Scholar] [CrossRef]
- Risi, C.M.; Belknap, B.; White, H.D.; Dryden, K.; Pinto, J.R.; Chase, P.B.; Galkin, V.E. High-resolution cryo-EM structure of the junction region of the native cardiac thin filament in relaxed state. PNAS Nexus 2023, 2, pgac298. [Google Scholar] [CrossRef]
- Risi, C.M.; Landim-Vieira, M.; Belknap, B.; Chase, P.B.; Pinto, J.R.; Galkin, V.E. The role of the troponin T interactions with actin in regulation of cardiac thin filament revealed by the troponin T pathogenic variant Ile79Asn. J. Mol. Cell. Cardiol. 2025, 204, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Namba, K.; Fujii, T. Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat. Commun. 2020, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Yanagisawa, H.; Wakabayashi, T. Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin. J. Struct. Biol. 2020, 209, 107450. [Google Scholar] [CrossRef]
- Ebashi, S.; Kodama, A. Interaction of troponin with F-actin in the presence of tropomyosin. J. Biochem. 1966, 59, 425–426. [Google Scholar] [CrossRef] [PubMed]
- Potter, J.D. The content of troponin, tropomyosin, actin, and myosin in rabbit skeletal muscle myofibrils. Arch. Biochem. Biophys. 1974, 162, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Potter, J.D.; Gergely, J. The calcium and magnesium binding sites on troponin and their role in the regulation of myofibrillar adenosine triphosphatase. J. Biol. Chem. 1975, 250, 4628–4633. [Google Scholar] [CrossRef] [PubMed]
- Isambert, H.; Venier, P.; Maggs, A.C.; Fattoum, A.; Kassab, R.; Pantaloni, D.; Carlier, M.-F. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995, 270, 11437–11444. [Google Scholar] [CrossRef]
- Loong, C.K.P.; Zhou, H.-X.; Chase, P.B. Persistence length of human cardiac α-tropomyosin measured by single molecule direct probe microscopy. PLoS ONE 2012, 7, e39676. [Google Scholar] [CrossRef]
- Bergmann, O.; Bhardwaj, R.D.; Bernard, S.; Zdunek, S.; Barnabé-Heider, F.; Walsh, S.; Zupicich, J.; Alkass, K.; Buchholz, B.A.; Druid, H.; et al. Evidence for cardiomyocyte renewal in humans. Science 2009, 324, 98–102. [Google Scholar] [CrossRef]
- Bergmann, O.; Zdunek, S.; Alkass, K.; Druid, H.; Bernard, S.; Frisén, J. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp. Cell Res. 2011, 317, 188–194. [Google Scholar] [CrossRef]
- Franklin, S.; Zhang, M.J.; Chen, H.; Paulsson, A.K.; Mitchell-Jordan, S.A.; Li, Y.; Ping, P.; Vondriska, T.M. Specialized compartments of cardiac nuclei exhibit distinct proteomic anatomy. Mol. Cell. Proteom. 2011, 10, M110.000703. [Google Scholar] [CrossRef]
- Ye, J.; Zhao, J.; Hoffmann-Rohrer, U.; Grummt, I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008, 22, 322–330. [Google Scholar] [CrossRef]
- Sahota, V.K.; Grau, B.F.; Mansilla, A.; Ferrús, A. Troponin I and Tropomyosin regulate chromosomal stability and cell polarity. J. Cell Sci. 2009, 122, 2623–2631. [Google Scholar] [CrossRef]
- Zhang, T.; Birbrair, A.; Delbono, O. Nonmyofilament-associated troponin T3 nuclear and nucleolar localization sequence and leucine zipper domain mediate muscle cell apoptosis. Cytoskeleton 2013, 70, 134–147. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Messi, M.L.; Pratley, R.E.; Zhang, T.; Delbono, O. Troponin T3 associates with DNA consensus sequence that overlaps with p53 binding motifs. Exp. Gerontol. 2018, 108, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Ljubojevic, S.; Bers, D.M. Nuclear calcium in cardiac myocytes. J. Cardiovasc. Pharmacol. 2015, 65, 211–217. [Google Scholar] [CrossRef]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef]
- Ljubojevic, S.; Radulovic, S.; Leitinger, G.; Sedej, S.; Sacherer, M.; Holzer, M.; Winkler, C.; Pritz, E.; Mittler, T.; Schmidt, A.; et al. Early remodeling of perinuclear Ca2+ stores and nucleoplasmic Ca2+ signaling during the development of hypertrophy and heart failure. Circulation 2014, 130, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.A.; Carrasco, M.A.; Adams, D.S.; Drouet, B.; Rios, J.; Müller, M.; Estrada, M.; Jaimovich, E. IP3 receptor function and localization in myotubes: An unexplored Ca2+ signaling pathway in skeletal muscle. J. Cell Sci. 2001, 114 Pt 20, 3673–3683. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.A.; Molgó, J.; Adams, D.S.; Colasante, C.; Williams, A.; Bohlen, M.; Jaimovich, E. IP3 receptors and associated Ca2+ signals localize to satellite cells and to components of the neuromuscular junction in skeletal muscle. J. Neurosci. 2003, 23, 8185–8192. [Google Scholar] [CrossRef]
- Ulferts, S.; Grosse, R. SUN2 mediates calcium-triggered nuclear actin polymerization to cluster active RNA polymerase II. EMBO Rep. 2024, 25, 4728–4748. [Google Scholar] [CrossRef]
- Dasgupta, I.; McCollum, D. Control of cellular responses to mechanical cues through YAP/TAZ regulation. J. Biol. Chem. 2019, 294, 17693–17706. [Google Scholar] [CrossRef]
- Fischer, M.; Rikeit, P.; Knaus, P.; Coirault, C. YAP-Mediated Mechanotransduction in Skeletal Muscle. Front. Physiol. 2016, 7, 41. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Kobirumaki-Shimozawa, F.; Inoue, T.; Shintani, S.A.; Oyama, K.; Terui, T.; Minamisawa, S.; Ishiwata, S.; Fukuda, N. Cardiac thin filament regulation and the Frank-Starling mechanism. J. Physiol. Sci. 2014, 64, 221–232. [Google Scholar] [CrossRef]
- Smith, L.; Tainter, C.; Regnier, M.; Martyn, D.A. Cooperative cross-bridge activation of thin filaments contributes to the Frank-Starling mechanism in cardiac muscle. Biophys. J. 2009, 96, 3692–3702. [Google Scholar] [CrossRef] [PubMed]
- Bensley, J.G.; De Matteo, R.; Harding, R.; Black, M.J. Three-dimensional direct measurement of cardiomyocyte volume, nuclearity, and ploidy in thick histological sections. Sci. Rep. 2016, 6, 23756. [Google Scholar] [CrossRef] [PubMed]
- Landim-Vieira, M.; Schipper, J.M.; Pinto, J.R.; Chase, P.B. Cardiomyocyte nuclearity and ploidy: When is double trouble? J. Muscle Res. Cell Motil. 2020, 41, 329–340. [Google Scholar] [CrossRef]
- Laflamme, M.A.; Murry, C.E. Heart regeneration. Nature 2011, 473, 326–335. [Google Scholar] [CrossRef]
- Jungbluth, H.; Gautel, M. Pathogenic mechanisms in centronuclear myopathies. Front. Aging Neurosci. 2014, 6, 339. [Google Scholar] [CrossRef]
- Mazzotti, A.L.; Coletti, D. The Need for a Consensus on the Locution “Central Nuclei” in Striated Muscle Myopathies. Front. Physiol. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.A.; Stroud, M.J. THE NUCLEUS: Mechanosensing in cardiac disease. Int. J. Biochem. Cell Biol. 2021, 137, 106035. [Google Scholar] [CrossRef]
- Coscarella, I.L.; Landim-Vieira, M.; Rastegarpouyani, H.; Chase, P.B.; Irianto, J.; Pinto, J.R. Nucleus mechanosensing in cardiomyocytes. Int. J. Mol. Sci. 2023, 24, 13341. [Google Scholar] [CrossRef]
- Landim-Vieira, M.; Nieto Morales, P.F.; ElSafty, S.; Kahmini, A.R.; Ranek, M.J.; Solís, C. The role of mechanosignaling in the control of myocardial mass. Am. J. Physiol. Heart Circ. Physiol. 2025, 328, H622–H638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Pereyra, A.S.; Wang, Z.-M.; Birbrair, A.; Reisz, J.A.; Files, D.C.; Purcell, L.; Feng, X.; Messi, M.L.; Feng, H.; et al. Calpain inhibition rescues troponin T3 fragmentation, increases Cav1.1, and enhances skeletal muscle force in aging sedentary mice. Aging Cell 2016, 15, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Huang, K.; Shi, M.; Huo, Y.; Han, L.; Liu, B.; Li, Y. Research Advances in the Role of the Tropomyosin Family in Cancer. Int. J. Mol. Sci. 2023, 24, 13295. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koopman, A.J.; Martin, A.J.; Moore, L.G.; Rodriguez, M.; Chase, P.B. RNA Polymerase II Activity and Nuclear Actin: Possible Roles of Nuclear Tropomyosin, Troponin and Ca2+ in Transcription in Striated Muscle Myocyte Nuclei. Macromol 2025, 5, 56. https://doi.org/10.3390/macromol5040056
Koopman AJ, Martin AJ, Moore LG, Rodriguez M, Chase PB. RNA Polymerase II Activity and Nuclear Actin: Possible Roles of Nuclear Tropomyosin, Troponin and Ca2+ in Transcription in Striated Muscle Myocyte Nuclei. Macromol. 2025; 5(4):56. https://doi.org/10.3390/macromol5040056
Chicago/Turabian StyleKoopman, Amelia J., Alexandra J. Martin, Lauren G. Moore, Michelle Rodriguez, and Prescott Bryant Chase. 2025. "RNA Polymerase II Activity and Nuclear Actin: Possible Roles of Nuclear Tropomyosin, Troponin and Ca2+ in Transcription in Striated Muscle Myocyte Nuclei" Macromol 5, no. 4: 56. https://doi.org/10.3390/macromol5040056
APA StyleKoopman, A. J., Martin, A. J., Moore, L. G., Rodriguez, M., & Chase, P. B. (2025). RNA Polymerase II Activity and Nuclear Actin: Possible Roles of Nuclear Tropomyosin, Troponin and Ca2+ in Transcription in Striated Muscle Myocyte Nuclei. Macromol, 5(4), 56. https://doi.org/10.3390/macromol5040056






