Predicting Tumor Progression in Patients with Cervical Cancer Using Computer Tomography Radiomic Features
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Image Preprocessing and Feature Determination
2.2. Data Cleaning and Transformation
2.3. Data Reduction
2.4. Model Building and Evaluation
3. Results
3.1. Patient Characteristics
3.2. Imaging Features Analysis
3.3. Classification Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRT | Concurrent chemo-radiotherapy |
| RT | Radiation Therapy |
| R | Recurrence |
| NR | Non-recurrence |
| CT | Computed Tomography |
| MRI | Magnetic Resonance Imaging |
| DW-MRI | Diffusion-Weighted Magnetic Resonance Imaging |
| PET | Positron Emission Tomography |
| GLCM | Gray-Level Co-Occurrence Matrix |
| GLDM | Gray-Level Dependence Matrix |
| SMOTE | Synthetic Minority Oversampling technique |
| LOOCV | Leave-One-Out Cross-Validation |
| GLRLM | Gray-Level Run Length Matrix |
| GLSZM | Gray-Level Size Zone Matrix |
| CV | Cross-Validation |
| ROI | Region of Interest |
| CI | Confidence Interval |
| LABC | Locally Advanced Breast Cancer |
| NAC | Neoadjuvant Chemotherapy |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Monk, B.J.; Tan, D.S.P.; Hernández Chagüi, J.D.; Takyar, J.; Paskow, M.J.; Nunes, A.T.; Pujade-Lauraine, E. Proportions and Incidence of Locally Advanced Cervical Cancer: A Global Systematic Literature Review. Int. J. Gynecol. Cancer 2022, 32, 1531–1539. [Google Scholar] [CrossRef] [PubMed]
- Datta, N.R.; Stutz, E.; Liu, M.; Rogers, S.; Klingbiel, D.; Siebenhüner, A.; Singh, S.; Bodis, S. Concurrent Chemoradiotherapy vs. Radiotherapy Alone in Locally Advanced Cervix Cancer: A Systematic Review and Meta-Analysis. Gynecol. Oncol. 2017, 145, 374–385. [Google Scholar] [CrossRef]
- Kawahara, D.; Nishibuchi, I.; Kawamura, M.; Yoshida, T.; Koh, I.; Tomono, K.; Sekine, M.; Takahashi, H.; Kikuchi, Y.; Kudo, Y.; et al. Radiomic Analysis for Pretreatment Prediction of Recurrence Post-Radiotherapy in Cervical Squamous Cell Carcinoma Cancer. Diagnostics 2022, 12, 2346. [Google Scholar] [CrossRef]
- Ren, K.; Shen, L.; Qiu, J.; Sun, K.; Chen, T.; Xuan, L.; Yang, M.; She, H.-Y.; Shen, L.; Zhu, H.; et al. Treatment Planning Computed Tomography Radiomics for Predicting Treatment Outcomes and Haematological Toxicities in Locally Advanced Cervical Cancer Treated with Radiotherapy: A Retrospective Cohort Study. BJOG 2023, 130, 222–230. [Google Scholar] [CrossRef]
- Park, H.; Kim, K.A.; Jung, J.-H.; Rhie, J.; Choi, S.Y. MRI Features and Texture Analysis for the Early Prediction of Therapeutic Response to Neoadjuvant Chemoradiotherapy and Tumor Recurrence of Locally Advanced Rectal Cancer. Eur. Radiol. 2020, 30, 4201–4211. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics 2019, 9, 1303–1322. [Google Scholar] [CrossRef] [PubMed]
- Ikushima, H.; Haga, A.; Ando, K.; Kato, S.; Kaneyasu, Y.; Uno, T.; Okonogi, N.; Yoshida, K.; Ariga, T.; Isohashi, F.; et al. Prediction of Out-of-Field Recurrence after Chemoradiotherapy for Cervical Cancer Using a Combination Model of Clinical Parameters and Magnetic Resonance Imaging Radiomics: A Multi-Institutional Study of the Japanese Radiation Oncology Study Group. J. Radiat. Res. 2022, 63, 98–106. [Google Scholar] [CrossRef]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and Limitations of Radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef]
- Verma, V.; Simone, C.B.; Krishnan, S.; Lin, S.H.; Yang, J.; Hahn, S.M. The Rise of Radiomics and Implications for Oncologic Management. J. Natl. Cancer Inst. 2017, 109, djx055. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Kan, Y.; Dong, D.; Yu, T.; Zhao, N.; Jiang, W.; Zhong, L.; Hu, C.; Luo, Y.; Tian, J. Multi-Habitat Based Radiomics for the Prediction of Treatment Response to Concurrent Chemotherapy and Radiation Therapy in Locally Advanced Cervical Cancer. Front. Oncol. 2020, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Bowen, S.R.; Yuh, W.T.C.; Hippe, D.S.; Wu, W.; Partridge, S.C.; Elias, S.; Jia, G.; Huang, Z.; Sandison, G.A.; Nelson, D.; et al. Tumor Radiomic Heterogeneity: Multiparametric Functional Imaging to Characterize Variability and Predict Response Following Cervical Cancer Radiation Therapy. J. Magn. Reson. Imaging 2018, 47, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Gui, B.; Autorino, R.; Miccò, M.; Nardangeli, A.; Pesce, A.; Lenkowicz, J.; Cusumano, D.; Russo, L.; Persiani, S.; Boldrini, L.; et al. Pretreatment MRI Radiomics Based Response Prediction Model in Locally Advanced Cervical Cancer. Diagnostics 2021, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, D.; Nishibuchi, I.; Kawamura, M.; Yoshida, T.; Nagata, Y. Radiomic Analysis for Pretreatment Prediction of Recurrence after Radiotherapy in Locally Advanced Cervical Cancer. Int. J. Radiat. Oncol. 2021, 111, e93. [Google Scholar] [CrossRef]
- Lucia, F.; Visvikis, D.; Vallières, M.; Desseroit, M.-C.; Miranda, O.; Robin, P.; Bonaffini, P.A.; Alfieri, J.; Masson, I.; Mervoyer, A.; et al. External Validation of a Combined PET and MRI Radiomics Model for Prediction of Recurrence in Cervical Cancer Patients Treated with Chemoradiotherapy. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 864–877. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Sharda, R.; Delen, D.T.E. Analytics, Data Sciences, & Artificial Intelligence: System for Decision Support, 11th ed.; Pearson: Harlow, UK, 2021. [Google Scholar]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Chu, A.; Sehgal, C.M.; Greenleaf, J.F. Use of Gray Value Distribution of Run Lengths for Texture Analysis. Pattern Recognit. Lett. 1990, 11, 415–419. [Google Scholar] [CrossRef]
- Tang, X. Texture Information in Run-Length Matrices. IEEE Trans. Image Process. 1998, 7, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M. Statistical and Structural Approaches to Texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Liu, F.T.; Ting, K.M.; Zhou, Z.-H. Isolation Forest. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining, Pisa, Italy, 15 December 2008; IEEE: Piscataway, NJ, USA, 2008; pp. 413–422. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-Sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2021, 45, 5–32. [Google Scholar] [CrossRef]
- Moghadas-Dastjerdi, H.; Sha-E-Tallat, H.R.; Sannachi, L.; Sadeghi-Naini, A.; Czarnota, G.J. A Priori Prediction of Tumour Response to Neoadjuvant Chemotherapy in Breast Cancer Patients Using Quantitative CT and Machine Learning. Sci. Rep. 2020, 10, 10936. [Google Scholar] [CrossRef]
- Moghadas-Dastjerdi, H.; Rahman, S.-E.-T.H.; Sannachi, L.; Wright, F.C.; Gandhi, S.; Trudeau, M.E.; Sadeghi-Naini, A.; Czarnota, G.J. Prediction of Chemotherapy Response in Breast Cancer Patients at Pre-Treatment Using Second Derivative Texture of CT Images and Machine Learning. Transl. Oncol. 2021, 14, 101183. [Google Scholar] [CrossRef]
- Reuzé, S.; Orlhac, F.; Chargari, C.; Nioche, C.; Limkin, E.; Riet, F.; Escande, A.; Haie-Meder, C.; Dercle, L.; Gouy, S.; et al. Prediction of Cervical Cancer Recurrence Using Textural Features Extracted from 18F-FDG PET Images Acquired with Different Scanners. Oncotarget 2017, 8, 43169–43179. [Google Scholar] [CrossRef]
- Lucia, F.; Visvikis, D.; Desseroit, M.-C.; Miranda, O.; Malhaire, J.-P.; Robin, P.; Pradier, O.; Hatt, M.; Schick, U. Prediction of Outcome Using Pretreatment 18F-FDG PET/CT and MRI Radiomics in Locally Advanced Cervical Cancer Treated with Chemoradiotherapy. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 768–786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, L.; Zhang, K.; Su, R.; Jia, H.; Qian, L.; Dong, J. Nomograms Combining Clinical and Imaging Parameters to Predict Recurrence and Disease-Free Survival After Concurrent Chemoradiotherapy in Patients with Locally Advanced Cervical Cancer. Acad. Radiol. 2023, 30, 499–508. [Google Scholar] [CrossRef]
- Zheng, R.; Cai, M.; Lan, L.; Huang, X.W.; Yang, Y.J.; Powell, M.; Lin, F. An MRI-Based Radiomics Signature and Clinical Characteristics for Survival Prediction in Early-Stage Cervical Cancer. Br. J. Radiol. 2022, 95, 20210838. [Google Scholar] [CrossRef]

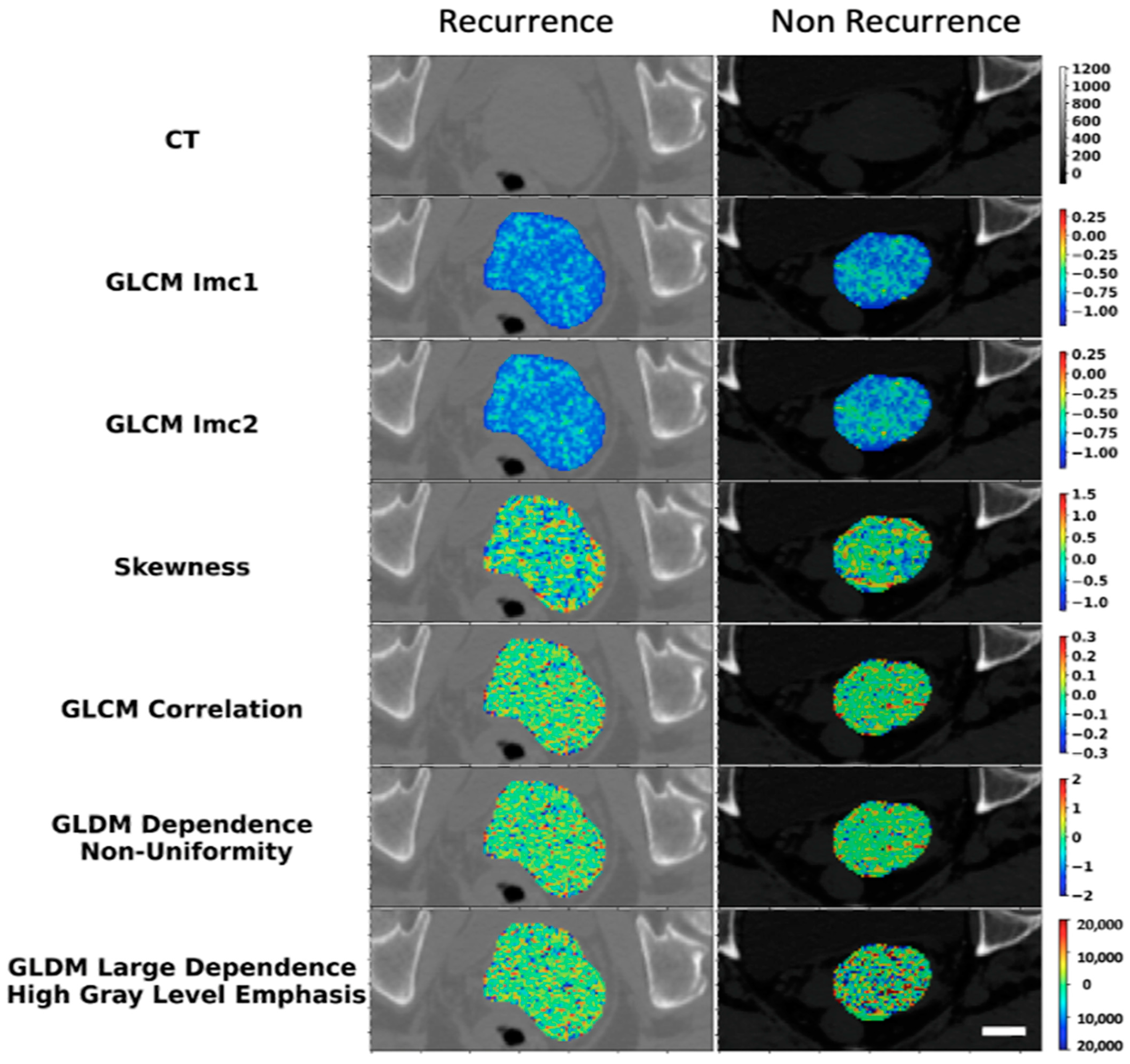
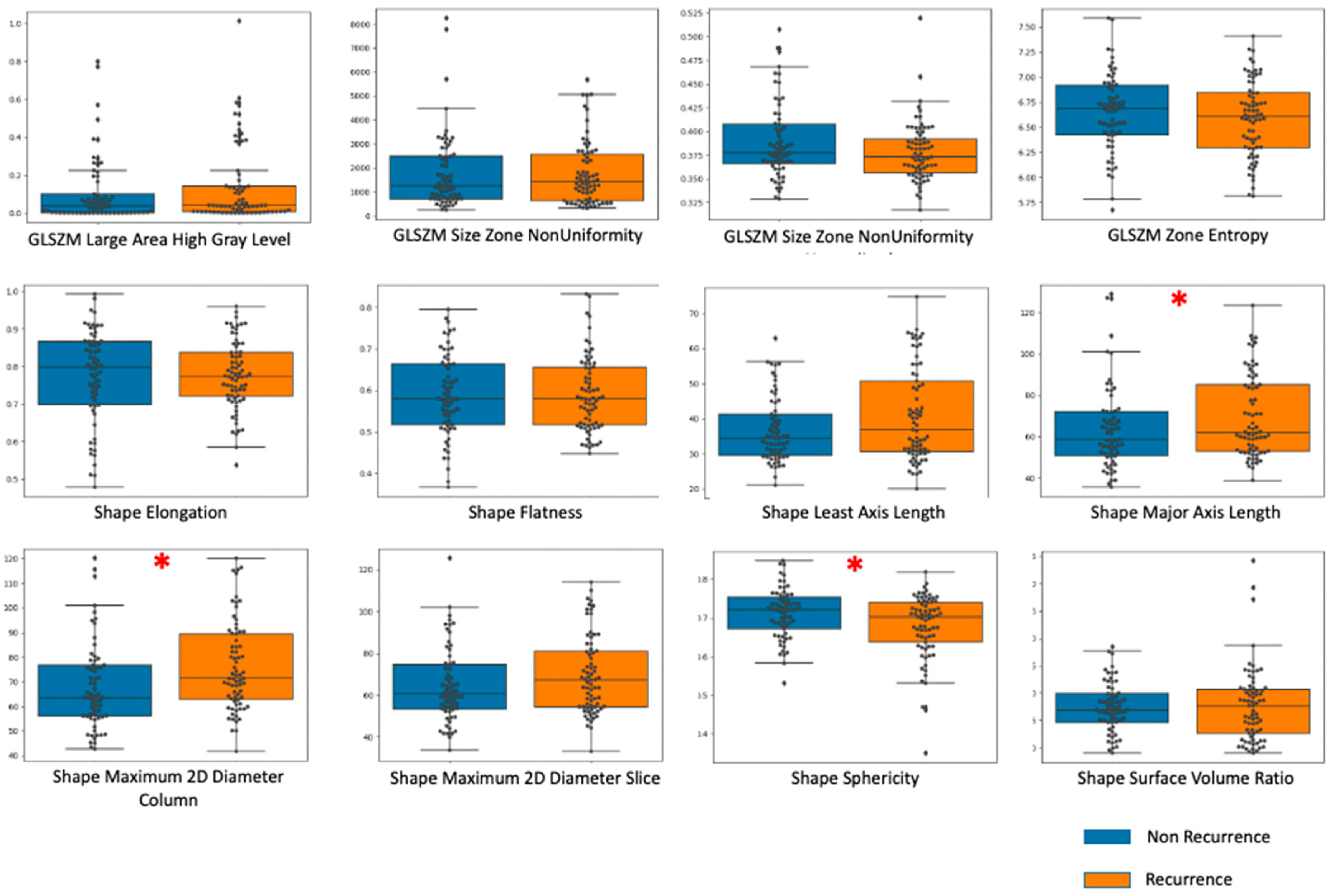

| Recurrence (n = 35) | Non-Recurrence (n = 77) | Total (n = 112) | |
|---|---|---|---|
| Age (Median, range) | 58 (33–81) | 57 (29–93) | 57 (33–93) |
| Primary Tumor Size (median) | 4.8 cm (2.0–8.2) | 3.9 cm (1.5–8.9) | 4.4 cm (1.5–8.9) |
| FIGO Staging of Cervical Cancer 2018 | |||
| IB | 2 (1.7%) | 22 (19.6%) | 24 (21.4%) |
| IIA | 1 (0.89%) | 2 (1.7%) | 3 (2.6%) |
| IIB | 10 (8.9%) | 40 (35.7%) | 50 (44.6%) |
| IIIA | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| IIIB | 4 (3.5%) | 2 (1.7%) | 6 (5.3%) |
| IIIC | 10 (8.9%) | 8 (7.1%) | 18 (16.0%) |
| IVA | 8 (7.1%) | 3 (2.6%) | 11 (9.8%) |
| Lymph Node Involvement | |||
| Pelvic | 13 (11.6%) | 10 (8.9%) | 23 (20.5%) |
| Para-aortic | 3 (2.6%) | 0 (0.0%) | 3 (2.6%) |
| Tumor Histology | |||
| Squamous Cell Carcinoma | 26 (23.2%) | 62 (55.3%) | 88 (78.5%) |
| Adenocarcinoma | 7 (6.2%) | 10 (8.9%) | 17 (15.1%) |
| Adenosquamous | 1 (1.7%) | 3 (2.6%) | 4 (3.5%) |
| Neuroendocrine | 1 (1.7%) | 2 (1.7%) | 3 (2.6%) |
| Histologic Tumor Grade | |||
| Well-differentiated | 0 (0.0%) | 6 (5.3%) | 6 (5.3%) |
| Moderately differentiated | 11 (9.8%) | 23 (20.5%) | 34 (30.3%) |
| Poorly differentiated | 14 (12.5%) | 21 (18.7%) | 35 (31.2%) |
| Not available | 10 (8.9%) | 27 (24.1%) | 37 (33.0%) |
| Radiation Dose | |||
| 4500 Gy in 25 fractions | 33 (29.4%) | 76 (67.8%) | 109 (97.3%) |
| 3000 Gy in 10 fractions * | 0 (0.0%) | 1 (0.89%) | 1 (0.8%) |
| 3750 Gy in 15 fractions | 1 (0.8%) | 0 (0.0%) | 1 (0.8%) |
| 5040 Gy in 28 fractions | 1 (0.8%) | 0 (0.0%) | 1 (0.8%) |
| Brachytherapy Dose | |||
| 2800 Gy in 4 fractions | 24 (21.4%) | 41 (36.6%) | 65 (58.0%) |
| 2400 Gy in 3 fractions | 7 (6.2%) | 25 (22.3%) | 32 (28.5%) |
| 2750 Gy in 5 fractions | 1 (0.8%) | 8 (7.1%) | 9 (8.0%) |
| 800 Gy in 1 fractions | 1 (0.8%) | 0 (0.0%) | 1 (0.8%) |
| 700 Gy in 1 fractions | 0 (0.0%) | 1 (0.8%) | 1 (0.8%) |
| 550 Gy in 1 fractions | 0 (0.0%) | 1 (0.8%) | 1 (0.8%) |
| None | 2 (1.7%) | 1 (0.8%) | 3 (2.6%) |
| Concurrent chemotherapy | |||
| Yes | 30 (26.7%) | 64 (57.1%) | 94 (83.9%) |
| No | 5 (4.4%) | 13 (11.6%) | 18 (16.0%) |
| Classifier | Sensitivity | Specificity | Precision | NPV | Accuracy | F1 Score | AUC |
|---|---|---|---|---|---|---|---|
| K-NN | 79.7 | 62.5 | 71.1 | 72.7 | 71.7 | 75.2 | 0.666 |
| RF | 79.7 | 75.0 | 78.7 | 76.2 | 77.5 | 79.2 | 0.784 |
| SVM Linear | 73.0 | 42.2 | 59.3 | 57.5 | 58.7 | 65.5 | 0.517 |
| SVM-RBF | 85.9 | 82.1 | 83.6 | 84.6 | 84.1 | 84.7 | 0.824 |
| Classifier | Sensitivity | Specificity | Precision | Accuracy | F1 Score | AUC |
|---|---|---|---|---|---|---|
| K-NN | 71.8 | 62.9 | 67.8 | 67.4 | 68.8 | 0.716 |
| RF | 71.4 | 68.3 | 74.1 | 70.4 | 71.5 | 0.740 |
| SVM Linear | 52.1 | 66.9 | 61.5 | 58.6 | 54.7 | 0.635 |
| SVM-RBF | 75.0 | 80.0 | 81.3 | 77.4 | 76.8 | 0.830 |
| Classifier | Sensitivity | Specificity | Precision | Accuracy | F1 Score | AUC |
|---|---|---|---|---|---|---|
| K-NN | 71.4 (3.87) | 57.2 (2.56) | 65.9 (1.96) | 64.7 (2.01) | 67.3 (2.61) | 0.689 (0.030) |
| RF | 69.3 (3.34) | 60.2 (3.20) | 67.8 (2.00) | 65.1 (1.90) | 67.6 (2.17) | 0.705 (0.018) |
| SVM Linear | 52.0 (3.17) | 58.0 (3.66) | 59.3 (3.07) | 54.7 (2.37) | 53.6 (2.62) | 0.563 (0.028) |
| SVM-RBF | 76.1 (1.47) | 70.4 (4.09) | 75.7 (2.81) | 73.6 (2.06) | 74.5 (1.72) | 0.794 (0.029) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasla, S.; Moore-Palhares, D.; Dicenzo, D.; Osapoetra, L.; Dasgupta, A.; Leung, E.; Barnes, E.; Hwang, A.; Taggar, A.S.; Czarnota, G.J. Predicting Tumor Progression in Patients with Cervical Cancer Using Computer Tomography Radiomic Features. Radiation 2024, 4, 355-368. https://doi.org/10.3390/radiation4040027
Prasla S, Moore-Palhares D, Dicenzo D, Osapoetra L, Dasgupta A, Leung E, Barnes E, Hwang A, Taggar AS, Czarnota GJ. Predicting Tumor Progression in Patients with Cervical Cancer Using Computer Tomography Radiomic Features. Radiation. 2024; 4(4):355-368. https://doi.org/10.3390/radiation4040027
Chicago/Turabian StylePrasla, Shopnil, Daniel Moore-Palhares, Daniel Dicenzo, LaurentiusOscar Osapoetra, Archya Dasgupta, Eric Leung, Elizabeth Barnes, Alexander Hwang, Amandeep S. Taggar, and Gregory Jan Czarnota. 2024. "Predicting Tumor Progression in Patients with Cervical Cancer Using Computer Tomography Radiomic Features" Radiation 4, no. 4: 355-368. https://doi.org/10.3390/radiation4040027
APA StylePrasla, S., Moore-Palhares, D., Dicenzo, D., Osapoetra, L., Dasgupta, A., Leung, E., Barnes, E., Hwang, A., Taggar, A. S., & Czarnota, G. J. (2024). Predicting Tumor Progression in Patients with Cervical Cancer Using Computer Tomography Radiomic Features. Radiation, 4(4), 355-368. https://doi.org/10.3390/radiation4040027







