Accurate Epigenetic Aging in Bottlenose Dolphins (Tursiops truncatus), an Essential Step in the Conservation of at-Risk Dolphins
Abstract
:1. Introduction
2. Materials and Methods
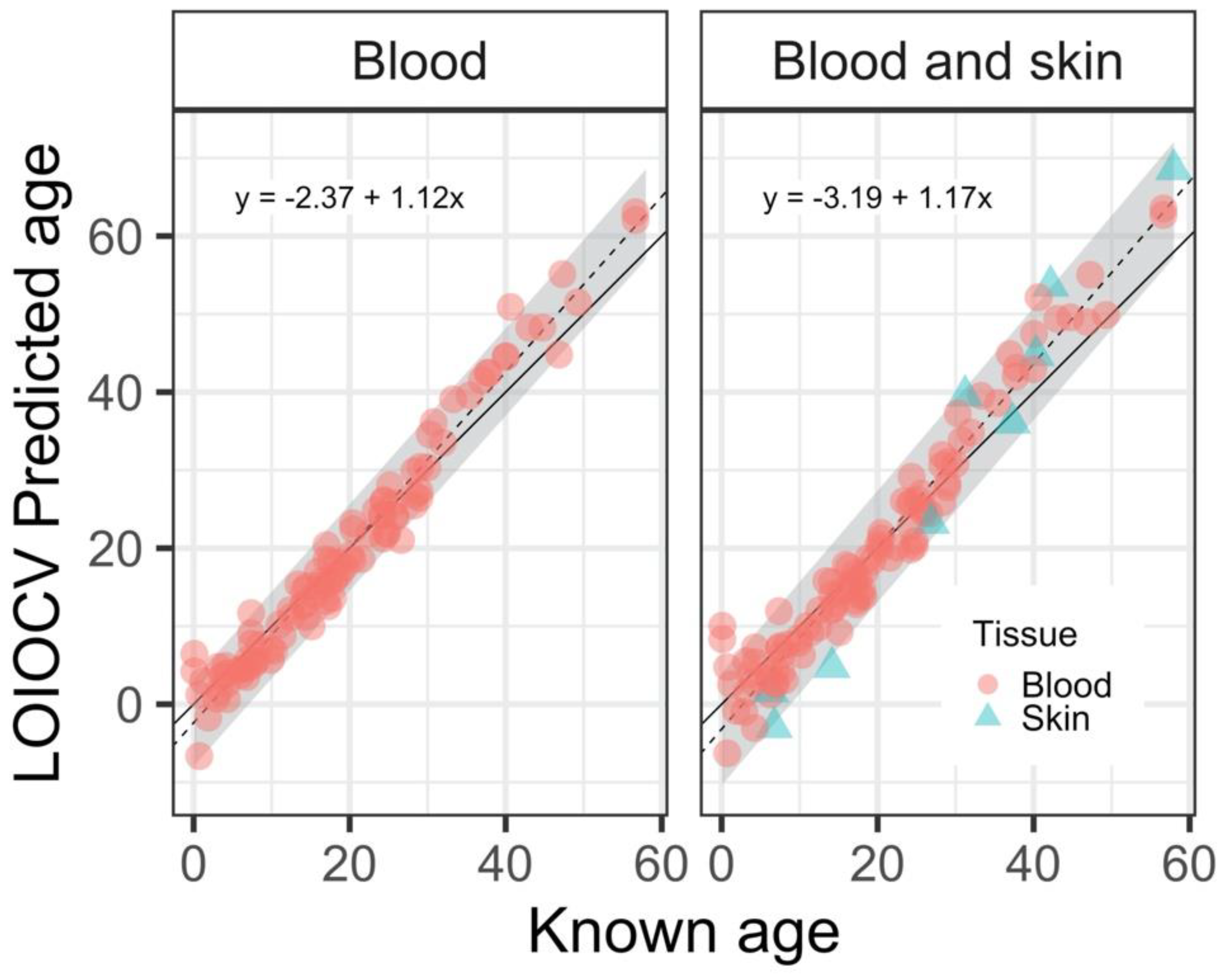
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barratclough, A.; Sanz-Requena, R.; Marti-Bonmati, L.; Schmitt, T.L.; Jensen, E.; García-Párraga, D. Radiographic assessment of pectoral flipper bone maturation in bottlenose dolphins (Tursiops truncatus), as a novel technique to accurately estimate chronological age. PLoS ONE 2019, 14, e0222722. [Google Scholar] [CrossRef] [PubMed]
- Hohn, A.A.; Fernandez, S. Biases in dolphin age structure due to age estimation technique. Mar. Mam. Sci. 1999, 15, 1124–1132. [Google Scholar] [CrossRef]
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sánchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic predictor of age. PLoS ONE 2011, 6, e14821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371. [Google Scholar] [CrossRef] [PubMed]
- Beal, A.; Kiszka, J.J.; Wells, R.S.; Eirin-Lopez, J.M. The Bottlenose dolphin Epigenetic Aging Tool (BEAT): A molecular age estimation tool for small cetaceans. Front. Mar. Sci. 2019, 6, 561. [Google Scholar] [CrossRef] [Green Version]
- Robeck, T.R.; Fei, Z.; Lu, A.T.; Haghani, A.; Jourdain, E.; Zoller, J.A.; Li, C.Z.; Steinman, K.J.; Di Rocco, S.; Schmitt, T.; et al. Multi-species and multi-tissue methylation clocks for age estimation in toothed whales and dolphins. Comm. Biol. 2021, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Paoli-Iseppi, D.; Deagle, B.E.; McMahon, C.R.; Hindell, M.A.; Dickinson, J.L.; Jarman, S.N. Measuring animal age with DNA methylation: From humans to wild animals. Front. Gen. 2017, 8, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridgway, S.H. Medical care of marine mammals. J. Am. Vet. Med. Ass. 1965, 147, 1077–1085. [Google Scholar]
- Bors, E.K.; Baker, C.S.; Wade, P.R.; O’Neill, K.B.; Shelden, K.E.; Thompson, M.J.; Fei, Z.; Jarman, S.; Horvath, S. An epigenetic clock to estimate the age of living beluga whales. Evol. App. 2020, 14, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Fahy, G.M.; Brooke, R.T.; Watson, J.P.; Good, Z.; Vasanawala, S.S.; Maecker, H.; Leipold, M.D.; Lin, D.T.; Kobor, M.S.; Horvath, S. Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell 2019, 18, e13028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arneson, A.; Haghani, A.; Thompson, M.J.; Pellegrini, M.; Kwon, S.B.; Vu, H.T.; Yao, M.; Li, C.Z.; Lu, A.T.; Barnes, B.; et al. A mammalian methylation array for profiling methylation levels at conserved sequences. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhou, W.; Triche Jr, T.J.; Laird, P.W.; Shen, H. SeSAMe: Reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018, 46, e123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B 2005, 67, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Soft. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Fasiolo, M.; Wood, S.N.; Zaffran, M.; Nedellec, R.; Goude, Y. qgam: Bayesian Non-Parametric Quantile Regression Modelling in R. Available online: https://arxiv.org/pdf/2007.03303.pdf (accessed on 26 May 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barratclough, A.; Smith, C.R.; Gomez, F.M.; Photopoulou, T.; Takeshita, R.; Pirotta, E.; Thomas, L.; McClain, A.M.; Parry, C.; Zoller, J.A.; et al. Accurate Epigenetic Aging in Bottlenose Dolphins (Tursiops truncatus), an Essential Step in the Conservation of at-Risk Dolphins. J. Zool. Bot. Gard. 2021, 2, 416-420. https://doi.org/10.3390/jzbg2030030
Barratclough A, Smith CR, Gomez FM, Photopoulou T, Takeshita R, Pirotta E, Thomas L, McClain AM, Parry C, Zoller JA, et al. Accurate Epigenetic Aging in Bottlenose Dolphins (Tursiops truncatus), an Essential Step in the Conservation of at-Risk Dolphins. Journal of Zoological and Botanical Gardens. 2021; 2(3):416-420. https://doi.org/10.3390/jzbg2030030
Chicago/Turabian StyleBarratclough, Ashley, Cynthia R. Smith, Forrest M. Gomez, Theoni Photopoulou, Ryan Takeshita, Enrico Pirotta, Len Thomas, Abby M. McClain, Celeste Parry, Joseph A. Zoller, and et al. 2021. "Accurate Epigenetic Aging in Bottlenose Dolphins (Tursiops truncatus), an Essential Step in the Conservation of at-Risk Dolphins" Journal of Zoological and Botanical Gardens 2, no. 3: 416-420. https://doi.org/10.3390/jzbg2030030
APA StyleBarratclough, A., Smith, C. R., Gomez, F. M., Photopoulou, T., Takeshita, R., Pirotta, E., Thomas, L., McClain, A. M., Parry, C., Zoller, J. A., Horvath, S., & Schwacke, L. H. (2021). Accurate Epigenetic Aging in Bottlenose Dolphins (Tursiops truncatus), an Essential Step in the Conservation of at-Risk Dolphins. Journal of Zoological and Botanical Gardens, 2(3), 416-420. https://doi.org/10.3390/jzbg2030030






