Immunological Mechanisms of Vaccine-Induced Protection against SARS-CoV-2 in Humans
Abstract
:1. Introduction
2. Sensing of SARS-CoV-2 Pathogen by Innate Immunity
3. Humoral and Cell Mediated Immune Responses against SARS-CoV-2
4. Vaccine-Induced Immune Responses against SARS-CoV-2 Infections
4.1. Nucleic Acid-Based Vaccines for COVID-19
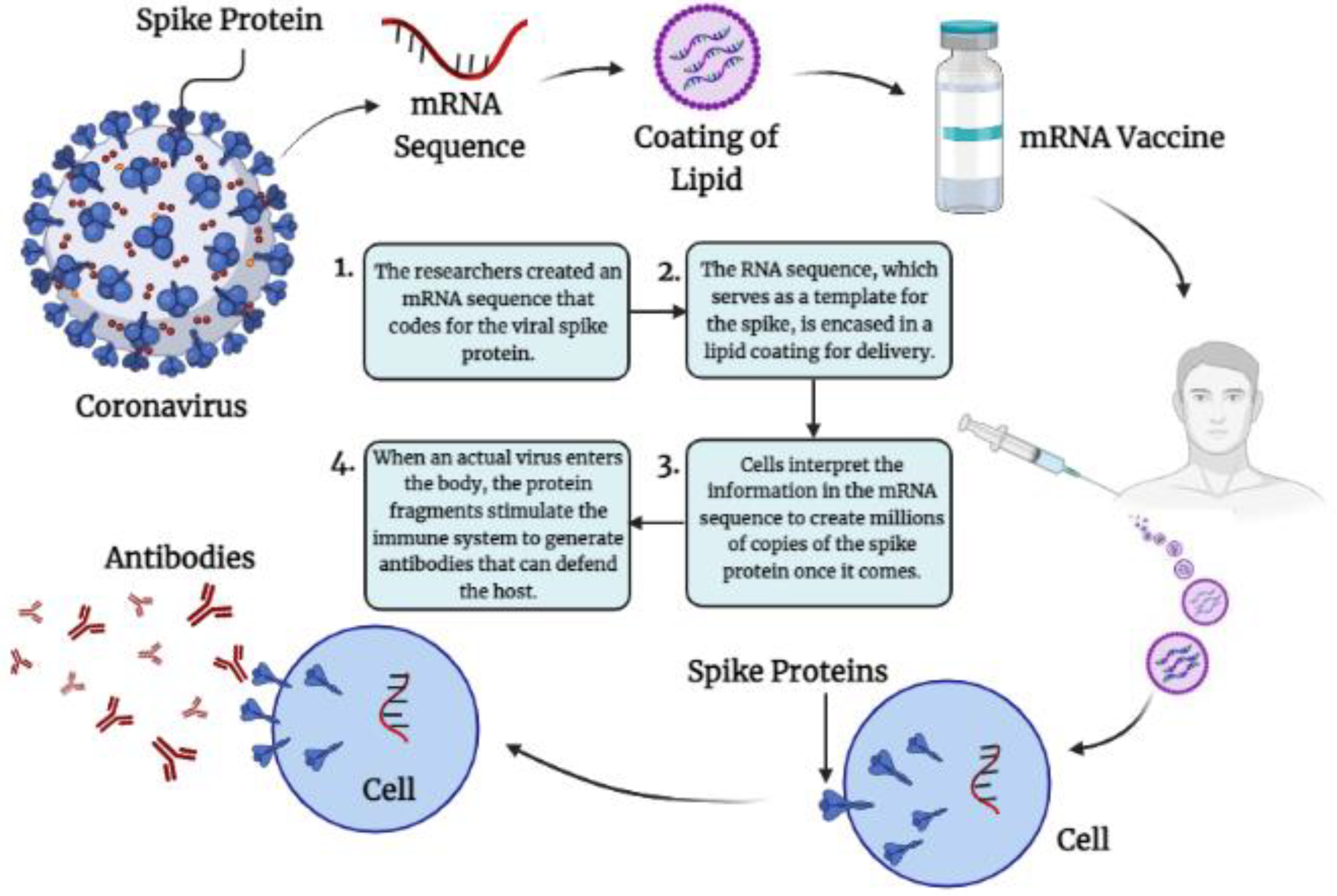
4.1.1. Immunological Mechanisms of Different m-RNA Vaccine-Induced Protection against SARS-CoV-2RNAVaccines
4.1.2. Immunological Mechanisms of Different DNA Based Vaccine-Induced Protection against SARS-CoV-2
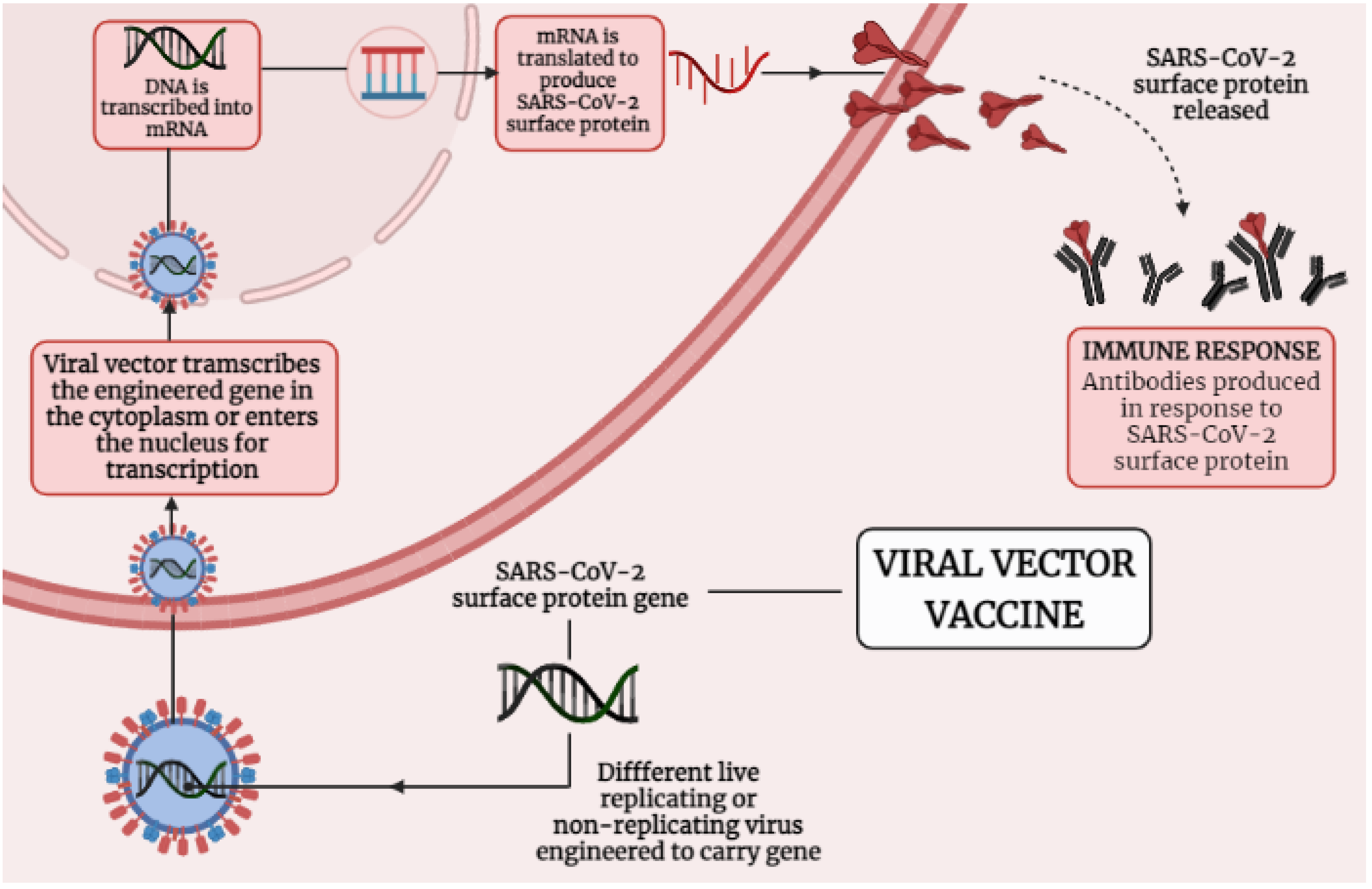
4.2. Immunological Mechanisms of Different Adenoviral Vector-Based Vaccines-Induced Protection against SARS-CoV-2
5. Vaccines and Its Role in Inducing Humoral Adaptive Immunity
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zeyaullah, M.; AlShahrani, A.M.; Muzammil, K.; Ahmad, I.; Alam, S.; Khan, W.H.; Ahmad, R. COVID-19 and SARS-CoV-2 Variants: Current Challenges and Health Concern. Front. Genet. 2021, 12, 693916. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 3 September 2021).
- Halstead, S.B.; Katzelnick, L. COVID-19 Vaccines: Should We Fear ADE? J. Infect. Dis. 2020, 222, 1946–1950. [Google Scholar] [CrossRef]
- Le, T.T.; Cramer, J.P.; Chen, R.; Mayhew, S. Evolution of the COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020, 19, 667–668. [Google Scholar] [CrossRef]
- Pronker, E.S.; Weenen, T.C.; Commandeur, H.; Claassen, E.H.; Osterhaus, A.D. Risk in vaccine research and development quantified. PLoS ONE 2013, 8, e57755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Godillot, A.P.; Madaio, M.P.; Weiner, D.B.; Williams, W.V. Vaccination against pathogenic cells by DNA inoculation. Curr. Top. Microbiol. Immunol. 1998, 226, 21–35. [Google Scholar] [PubMed]
- Davis, H.L.; Demeneix, B.A.; Quantin, B.; Coulombe, J.; Whalen, R.G. Plasmid DNA is superior to viral vectors for direct gene transfer into adult mouse skeletal muscle. Hum. Gene Ther. 1993, 4, 733–740. [Google Scholar] [CrossRef]
- Hurpin, C.; Rotarioa, C.; Bisceglia, H.; Chevalier, M.; Tartaglia, J.; Erdile, L. The mode of presentation and route of administration are critical for the induction of immune responses to p53 and antitumor immunity. Vaccine 1998, 16, 208–215. [Google Scholar] [CrossRef]
- Sadarangani, M.; Marchant, A.; Kollmann, T.R. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 2021, 21, 475–484. [Google Scholar] [CrossRef] [PubMed]
- The Adaptive Immune System. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. Available online: https://www.ncbi.nlm.nih.gov/books/NBK21070/ (accessed on 2 September 2021).
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulendran, B. Modulating vaccine responses with dendritic cells and Toll-like receptors. Immunol. Rev. 2004, 199, 227–250. [Google Scholar] [CrossRef]
- Loo, Y.M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; García-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Takeshita, F.; Kobiyama, K.; Miyawaki, A.; Jounai, N.; Okuda, K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy 2008, 4, 67–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Young, B.E.; Fong, S.W.; Chan, Y.H.; Mak, T.M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 2020, 396, 603–611. [Google Scholar] [CrossRef]
- Gudbjartsson, D.F.; Norddahl, G.L.; Melsted, P.; Gunnarsdottir, K.; Holm, H.; Eythorsson, E.; Arnthorsson, A.O.; Helgason, D.; Bjarnadottir, K.; Ingvarsson, R.F.; et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020, 383, 1724–1734. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [Green Version]
- The Humoral Immune Response. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. Available online: https://www.ncbi.nlm.nih.gov/books/NBK10752/ (accessed on 1 September 2021).
- Jordan, S.C. Innate and adaptive immune responses to SARS-CoV-2 in humans: Relevance to acquired immunity and vaccine responses. Clin. Exp. Immunol. 2021, 204, 310–320. [Google Scholar] [CrossRef]
- Stephens, D.S.; McElrath, M.J. COVID-19 and the Path to Immunity. JAMA 2020, 324, 1279–1281. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Zimmerman, M.G.; Kauffman, R.C.; Mantus, G.; Linderman, S.L.; Hudson, W.H.; Vanderheiden, A.; Nyhoff, L.; Davis, C.W.; Adekunle, O.; et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corr, M.; Lee, D.J.; Carson, D.A.; Tighe, H. Gene vaccination with naked plasmid DNA: Mechanism of CTL priming. J. Exp. Med. 1996, 184, 1555–1560. [Google Scholar] [CrossRef] [PubMed]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Porgador, A.; Irvine, K.R.; Iwasaki, A.; Barber, B.H.; Restifo, N.P.; Germain, R.N. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. J. Exp. Med. 1998, 188, 1075–1082. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Bettini, E.; Locci, M. SARS-CoV-2 mRNA Vaccines: Immunological Mechanism and Beyond. Vaccines 2021, 9, 147. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Hobo, W.; Novobrantseva, T.I.; Fredrix, H.; Wong, J.; Milstein, S.; Epstein-Barash, H.; Liu, J.; Schaap, N.; van der Voort, R.; Dolstra, H. Improving dendritic cell vaccine immunogenicity by silencing PD-1 ligands using siRNA-lipid nanoparticles combined with antigen mRNA electroporation. Cancer Immunol. Immunother. 2013, 62, 285–297. [Google Scholar] [CrossRef]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef]
- Martinon, F.; Krishnan, S.; Lenzen, G.; Magné, R.; Gomard, E.; Guillet, J.G.; Lévy, J.P.; Meulien, P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993, 23, 1719–1722. [Google Scholar] [CrossRef]
- Pollard, C.; De Koker, S.; Saelens, X.; Vanham, G.; Grooten, J. Challenges and advances towards the rational design of mRNA vaccines. Trends Mol. Med. 2013, 19, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, K.E.; Bhosle, S.M.; Zurla, C.; Beyersdorf, J.; Rogers, K.A.; Vanover, D.; Xiao, P.; Araínga, M.; Shirreff, L.M.; Pitard, B.; et al. Visualization of early events in mRNA vaccine delivery in non-human primates via PET-CT and near-infrared imaging. Nat. Biomed. Eng. 2019, 3, 371–380. [Google Scholar] [CrossRef]
- Pepini, T.; Pulichino, A.M.; Carsillo, T.; Carlson, A.L.; Sari-Sarraf, F.; Ramsauer, K.; Debasitis, J.C.; Maruggi, G.; Otten, G.R.; Geall, A.J.; et al. Induction of an IFN-Mediated Antiviral Response by a Self-Amplifying RNA Vaccine: Implications for Vaccine Design. J. Immunol. 2017, 198, 4012–4024. [Google Scholar] [CrossRef] [Green Version]
- Drugs and Lactation Database (LactMed) [Internet]. Bethesda (MD): National Library of Medicine (US); 2006-COVID-19 Vaccines. [Updated 2021 August 16]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK565969/ (accessed on 4 September 2021).
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Zeng, J.; Yan, J. COVID-19 mRNA vaccines. J. Genet. Genom. 2021, 48, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Silveira, M.M.; Moreira, G.; Mendonça, M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef] [PubMed]
- Comirnaty: COVID-19 mRNA Vaccine (Nucleoside-Modified). Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/comirnaty (accessed on 3 September 2021).
- Comirnaty: CHMP Public Assessment Report. Available online: https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar-public-assessment-report_en.pdf (accessed on 6 September 2021).
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. C4591001 Clinical Trial Group. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R., Jr.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Borah, P.; Deb, P.K.; Al-Shar’i, N.A.; Dahabiyeh, L.A.; Venugopala, K.N.; Singh, V.; Shinu, P.; Hussain, S.; Deka, S.; Chandrasekaran, B.; et al. Perspectives on RNA Vaccine Candidates for COVID-19. Front. Mol. Biosci. 2021, 8, 635245. [Google Scholar] [CrossRef]
- Rauch, S.; Roth, N.; Schwendt, K.; Fotin-Mleczek, M.; Mueller, S.O.; Petsch, B. mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. NPJ Vaccines 2021, 6, 57. [Google Scholar] [CrossRef]
- Zhang, N.N.; Li, X.F.; Deng, Y.Q.; Zhao, H.; Huang, Y.J.; Yang, G.; Huang, W.J.; Gao, P.; Zhou, C.; Zhang, R.R.; et al. A Thermostable mRNA Vaccine against COVID-19. Cell 2020, 182, 1271–1283. [Google Scholar] [CrossRef]
- Rawat, K.; Kumari, P.; Saha, L. COVID-19 vaccine: A recent update in pipeline vaccines, their design and development strategies. Eur. J. Pharmacol. 2021, 892, 173751. [Google Scholar] [CrossRef]
- de Queiroz, N.; Marinho, F.V.; Chagas, M.A.; Leite, L.; Homan, E.J.; de Magalhães, M.; Oliveira, S.C. Vaccines for COVID-19: Perspectives from nucleic acid vaccines to BCG as delivery vector system. Microbes Infect. 2020, 22, 515–524. [Google Scholar] [CrossRef]
- Leitner, W.W.; Ying, H.; Restifo, N.P. DNA and RNA-based vaccines: Principles, progress and prospects. Vaccine 1999, 18, 765–777. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.C.; DeVit, M.; Johnston, S.A. Genetic immunization is a simple method for eliciting an immune response. Nature 1992, 356, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.A. DNA vaccines: An historical perspective and view to the future. Immunol. Rev. 2011, 239, 62–84. [Google Scholar] [CrossRef]
- Schirmbeck, R.; Böhm, W.; Reimann, J. DNA vaccination primes MHC class I-restricted, simian virus 40 large tumor antigen-specific CTL in H-2d mice that reject syngeneic tumors. J. Immunol. 1996, 157, 3550–3558. [Google Scholar] [PubMed]
- Silveira, M.M.; Oliveira, T.L.; Schuch, R.A.; McBride, A.; Dellagostin, O.A.; Hartwig, D.D. DNA vaccines against leptospirosis: A literature review. Vaccine 2017, 35, 5559–5567. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Izzard, L.; Hurt, A.C. A Review of DNA Vaccines Against Influenza. Front. Immunol. 2018, 9, 1568. [Google Scholar] [CrossRef] [Green Version]
- Coban, C.; Kobiyama, K.; Jounai, N.; Tozuka, M.; Ishii, K.J. DNA vaccines: A simple DNA sensing matter? Hum. Vaccines Immunother. 2013, 9, 2216–2221. [Google Scholar] [CrossRef] [Green Version]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef]
- Funk, C.D.; Laferrière, C.; Ardakani, A. A Snapshot of the Global Race for Vaccines Targeting SARS-CoV-2 and the COVID-19 Pandemic. Front. Pharmacol. 2020, 11, 937. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.B.; Suh, Y.S.; Ryu, J.I.; Jang, H.; Oh, H.; Koo, B.S.; Seo, S.H.; Hong, J.J.; Song, M.; Kim, S.J.; et al. Soluble Spike DNA Vaccine Provides Long-Term Protective Immunity against SARS-CoV-2 in Mice and Nonhuman Primates. Vaccines 2021, 9, 307. [Google Scholar] [CrossRef]
- Ramasamy, M.N.; Minassian, A.M.; Ewer, K.J.; Flaxman, A.L.; Folegatti, P.M.; Owens, D.R.; Voysey, M.; Aley, P.K.; Angus, B.; Babbage, G.; et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 2021, 396, 1979–1993. [Google Scholar] [CrossRef]
- Rayburn, E.R.; Zhang, R. Antisense, RNAi, and gene silencing strategies for therapy: Mission possible or impossible? Drug Discov. Today 2008, 13, 513–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akita, H.; Kogure, K.; Moriguchi, R.; Nakamura, Y.; Higashi, T.; Nakamura, T.; Serada, S.; Fujimoto, M.; Naka, T.; Futaki, S.; et al. Nanoparticles for ex vivo siRNA delivery to dendritic cells for cancer vaccines: Programmed endosomal escape and dissociation. J. Control. Release Off. J. Control. Release Soc. 2010, 143, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Piggott, J.M.; Sheahan, B.J.; Soden, D.M.; O’Sullivan, G.C.; Atkins, G.J. Electroporation of RNA stimulates immunity to an encoded reporter gene in mice. Mol. Med. Rep. 2009, 2, 753–756. [Google Scholar]
- Diken, M.; Kreiter, S.; Selmi, A.; Türeci, O.; Sahin, U. Antitumor vaccination with synthetic mRNA: Strategies for in vitro and in vivo preclinical studies. Methods Mol. Biol. 2013, 969, 235–246. [Google Scholar]
- Pollard, C.; Rejman, J.; De Haes, W.; Verrier, B.; Van Gulck, E.; Naessens, T.; De Smedt, S.; Bogaert, P.; Grooten, J.; Vanham, G.; et al. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Weiss, R.; Scheiblhofer, S.; Roesler, E.; Weinberger, E.; Thalhamer, J. mRNA vaccination as a safe approach for specific protection from type I allergy. Expert Rev. Vaccines 2012, 11, 55–67. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.; Sarkar, S.; Gourley, T.S.; Rouse, B.T.; Ahmed, R. Differentiation of memory B and T cells. Curr. Opin. 2006, 18, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Quan, F.S.; Huang, C.; Compans, R.W.; Kang, S.M. Virus-like particle vaccine induces protective immunity against homologous and heterologous strains of influenza virus. J. Virol. 2007, 81, 3514–3524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, F.S.; Compans, R.W.; Nguyen, H.H.; Kang, S.M. Induction of heterosubtypic immunity to influenza virus by intranasal immunization. J. Virol. 2008, 82, 1350–1359. [Google Scholar] [CrossRef] [Green Version]
- Crotty, S.; Aubert, R.D.; Glidewell, J.; Ahmed, R. Tracking human antigen-specific memory B cells: A sensitive and generalized ELISPOT system. J. Immunol. Methods 2004, 286, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 2007, 25, 1361–1366. [Google Scholar] [CrossRef]
- Sancho, D.; Gómez, M.; Sánchez-Madrid, F. CD69 is an immunoregulatory molecule induced following activation. Trends Immunol. 2005, 26, 136–140. [Google Scholar] [CrossRef] [PubMed]
- McHeyzer-Williams, L.J.; McHeyzer-Williams, M.G. Antigen-specific memory B cell development. Annu. Rev. Immunol. 2005, 23, 487–513. [Google Scholar] [CrossRef] [PubMed]
- Goel, H.; Gupta, I.; Mourya, M.; Gill, S.; Chopra, A.; Ranjan, A.; Rath, G.K.; Tanwar, P.A. systematic review of clinical and laboratory parameters of 3000 COVID-19 cases. Obstet. Gynecol. Sci. 2021, 64, 174–189. [Google Scholar] [CrossRef]
- Reif, K.; Ekland, E.H.; Ohl, L.; Nakano, H.; Lipp, M.; Förster, R.; Cyster, J.G. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature 2002, 416, 94–99. [Google Scholar] [CrossRef]
- Kelsoe, G. Studies of the humoral immune response. Immunol. Res. 2000, 22, 199–210. [Google Scholar] [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Goel, H.; Goyal, K.; Baranwal, P.; Dixit, A.; Upadhyay, T.K.; Upadhye, V.J. The Diagnostics Technologies and Control of COVID-19. Lett. Appl. NanoBioSci. 2021, 11, 3120–3133. [Google Scholar]
- Secchi, M.; Bazzigaluppi, E.; Brigatti, C.; Marzinotto, I.; Tresoldi, C.; Rovere-Querini, P.; Poli, A.; Castagna, A.; Scarlatti, G.; Zangrillo, A.; et al. COVID-19 survival associates with the immunoglobulin response to the SARS-CoV-2 spike receptor binding domain. J. Clin. Investig. 2020, 130, 6366–6378. [Google Scholar] [CrossRef]

| Covishield | Covaxin | |
|---|---|---|
| DEVELOPED BY | Serum Institute of India | Bharat Biotech ICMR |
| VACCINE TYPE | Non-Replicating Viral Vector | Inactivated |
| EFFICACY | Drugs Controller General of India (DCGI): 70.42% overall | 60% |
| STORAGE TEMPERATURE | 2–8 degree Celsius | 2–8 degree Celsius |
| DOSES | Two Doses | Two Doses |
| (0, 84 Days) | (0, 14 Days) |
| Vaccine Developed by | Name of Vaccine | Mode | Type of Response |
|---|---|---|---|
| Pfizer/BioNtech | BNT162b2 | mRNA vaccine | IgG, IgA, CD8+ cells or CD4+ cells |
| Moderna | mRNA-1273 | mRNA vaccine | CD8+ T cell |
| CureVac AG | CVnCoV | mRNA vaccine | IL-6, IFN-α |
| Abogen | ARCoV | mRNA vaccine | Th-1 biased |
| Arcturus | ARCT-021 | mRNA vaccine | CD8+ cell-mediated and Th1/Th2-mediated immunity |
| Symvivo | BacTRL-Spike | DNA vaccine | Induce both cellular and humoral immunity against spike protein |
| Genexine | GX-19 | DNA vaccine | Th1-biased T cell responses, CD4+ and CD8+ T cell |
| Inovio | INO-4800 | DNA vaccine | IgG cell |
| AnGes Inc. and Osaka University | AG0301-COVID19 | DNA vaccine | Neutralizing antibodies and T-cell responses |
| Inovio | GLS-5300 | DNA vaccine | T-cell responses, S1-ELISA |
| Oxford/AstraZeneca | ChAdOx1 nCoV-19 | Adenoviral-vectored | Anti-IgA and IgG antibodies, T cell, Th1-biased T-cell, IFN-γ and IL-2, CD4+ T cells |
| Gamaleya Research Institute | Gam-COVID-Vac (Sputnik V) | Adenoviral-vectored | IgG cell |
| Johnson and Johnson | Janssen, Ad26.COV2.S | Adenoviral-vectored | Th1-biased, Th2-skewed, CD8+ T-cell, IFN-γ, IL-4, IL-5, or IL-10 |
| Bharat Biotech | BBV152 | Whole cell inactivated viral vaccine | Th-1 cells, IgG cells |
| Sinovac Biotech | SinoVec | Inactivated-virus COVID-19 Vaccine | T cells |
| Beijing Bio-Institute of Biological Products Co Ltd. | Sinopharm | Inactivated-virus COVID-19 Vaccine | Neutralizing antibody GMT, Humoral responses |
| CanSino Biologics Inc. | CanSino | Inactivated-virus COVID-19 vaccine | Specific ELISA antibody responses to the receptor binding domain (RBD) and neutralizing antibody responses |
| Novavex | NVX-CoV2372 | Protein subunit vaccine | CD4+ T-cell, IgG cells |
| Vektor State Research Center of Virology and Biotechnology in Russia | EpiVecCorona | Protein subunit vaccine | CD4+ T-cell |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goyal, K.; Goel, H.; Baranwal, P.; Tewary, A.; Dixit, A.; Pandey, A.K.; Benjamin, M.; Tanwar, P.; Dey, A.; Khan, F.; et al. Immunological Mechanisms of Vaccine-Induced Protection against SARS-CoV-2 in Humans. Immuno 2021, 1, 442-456. https://doi.org/10.3390/immuno1040032
Goyal K, Goel H, Baranwal P, Tewary A, Dixit A, Pandey AK, Benjamin M, Tanwar P, Dey A, Khan F, et al. Immunological Mechanisms of Vaccine-Induced Protection against SARS-CoV-2 in Humans. Immuno. 2021; 1(4):442-456. https://doi.org/10.3390/immuno1040032
Chicago/Turabian StyleGoyal, Keshav, Harsh Goel, Pritika Baranwal, Anisha Tewary, Aman Dixit, Avanish Kumar Pandey, Mercilena Benjamin, Pranay Tanwar, Abhijit Dey, Fahad Khan, and et al. 2021. "Immunological Mechanisms of Vaccine-Induced Protection against SARS-CoV-2 in Humans" Immuno 1, no. 4: 442-456. https://doi.org/10.3390/immuno1040032
APA StyleGoyal, K., Goel, H., Baranwal, P., Tewary, A., Dixit, A., Pandey, A. K., Benjamin, M., Tanwar, P., Dey, A., Khan, F., Pandey, P., Gupta, P. K., Kumar, D., Roychoudhury, S., Jha, N. K., Upadhyay, T. K., & Kesari, K. K. (2021). Immunological Mechanisms of Vaccine-Induced Protection against SARS-CoV-2 in Humans. Immuno, 1(4), 442-456. https://doi.org/10.3390/immuno1040032












