An Altered Gut Microbiota–Brain Axis in Fragile X Syndrome May Explain Autistic Traits in Some Patients
Abstract
1. Introduction
1.1. Fragile X Syndrome, Autism Spectrum Disorder, and Their Relationship with Gut Microbiota
1.1.1. Fragile X Syndrome
1.1.2. Microbiome Alterations in Autism Spectrum Disorders
1.2. Human Gut Microbiota: Factors Influencing Its Composition
1.3. Relationship Between Gut Microbiota and the Nervous System: The Gut–Brain Axis
1.4. Gut Microbiota Influence on Brain Development and Function
1.5. Influence of Gut Microbiota on Hippocampal Neurogenesis and the Blood–Brain Barrier
2. Materials and Methods
2.1. Study Setting and Population
2.1.1. Eligibility Criteria
2.1.2. Age Range Considerations
2.1.3. Ethical Considerations
2.2. Collection, Transport, and Processing of Biological Samples
2.3. DNA Extraction
2.4. 16S rRNA Gene-Based Sequencing
Sequencing Steps
2.5. Read Assembly and Quality Control
2.6. Taxonomic Annotation
2.6.1. Operational Taxonomic Unit (OTU) Classification
2.6.2. Taxonomic Identification
2.6.3. Phylogenetic and Abundance Analysis
2.7. Assessment of Read Coverage and Alpha and Beta Diversity Calculation
2.7.1. Alpha Diversity
Species Richness
Statistical Analysis of Alpha Diversity
2.7.2. Beta Diversity
Taxonomic Differential Abundance Analysis
LEfSe Analysis (Linear Discriminant Analysis Effect Size)
3. Results
3.1. Alpha Diversity
3.2. Gut Microbiota Composition
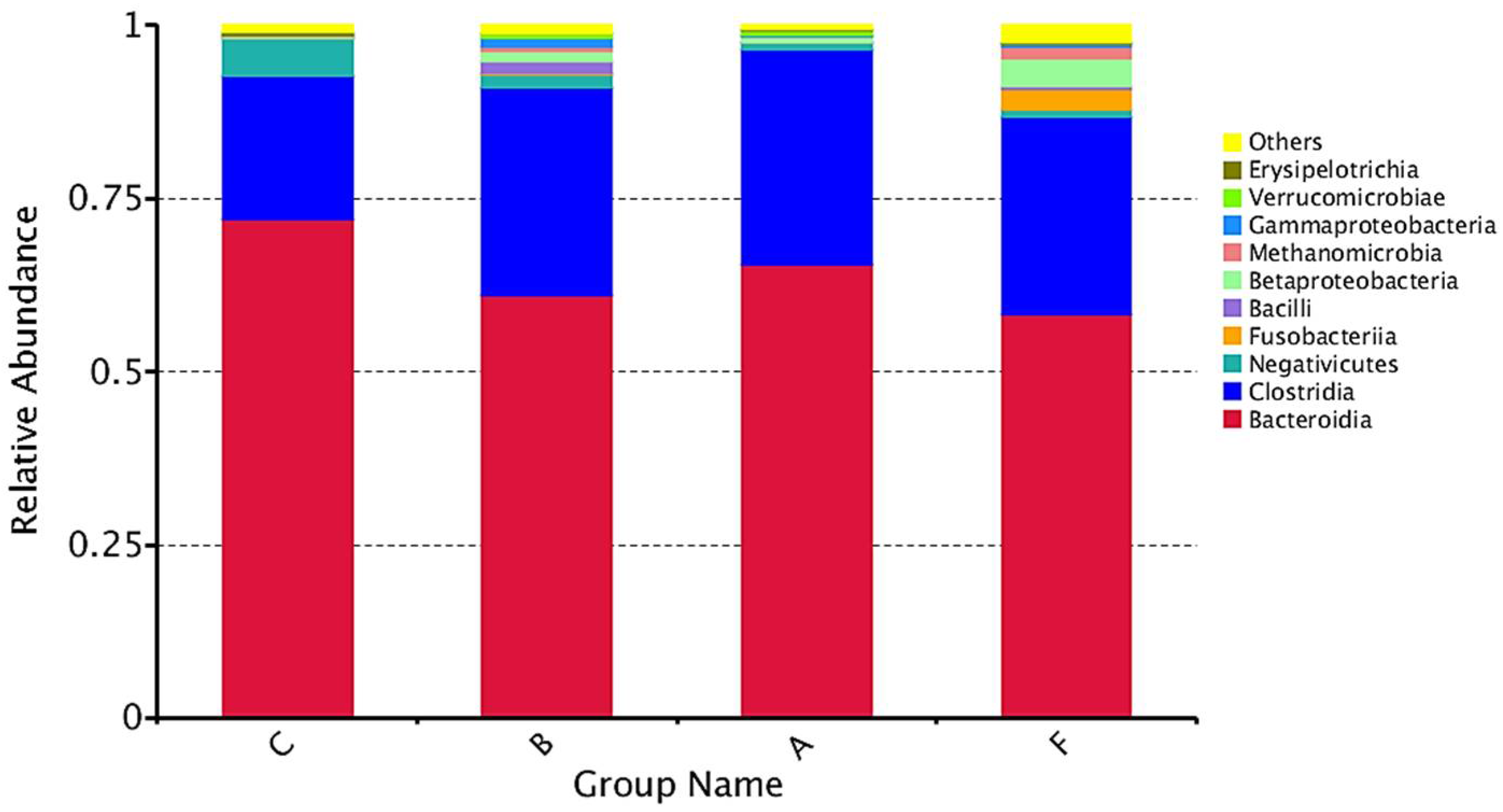
3.2.1. Phylum-Level Analysis
3.2.2. Class-Level Analysis
3.2.3. Heatmap Analysis
3.3. Beta Diversity
3.4. Biomarker Analysis Through Linear Discriminant Analysis (LDA)
4. Discussion
4.1. Alpha Diversity
4.2. Microbiota Composition
4.3. Heatmap and Beta Diversity
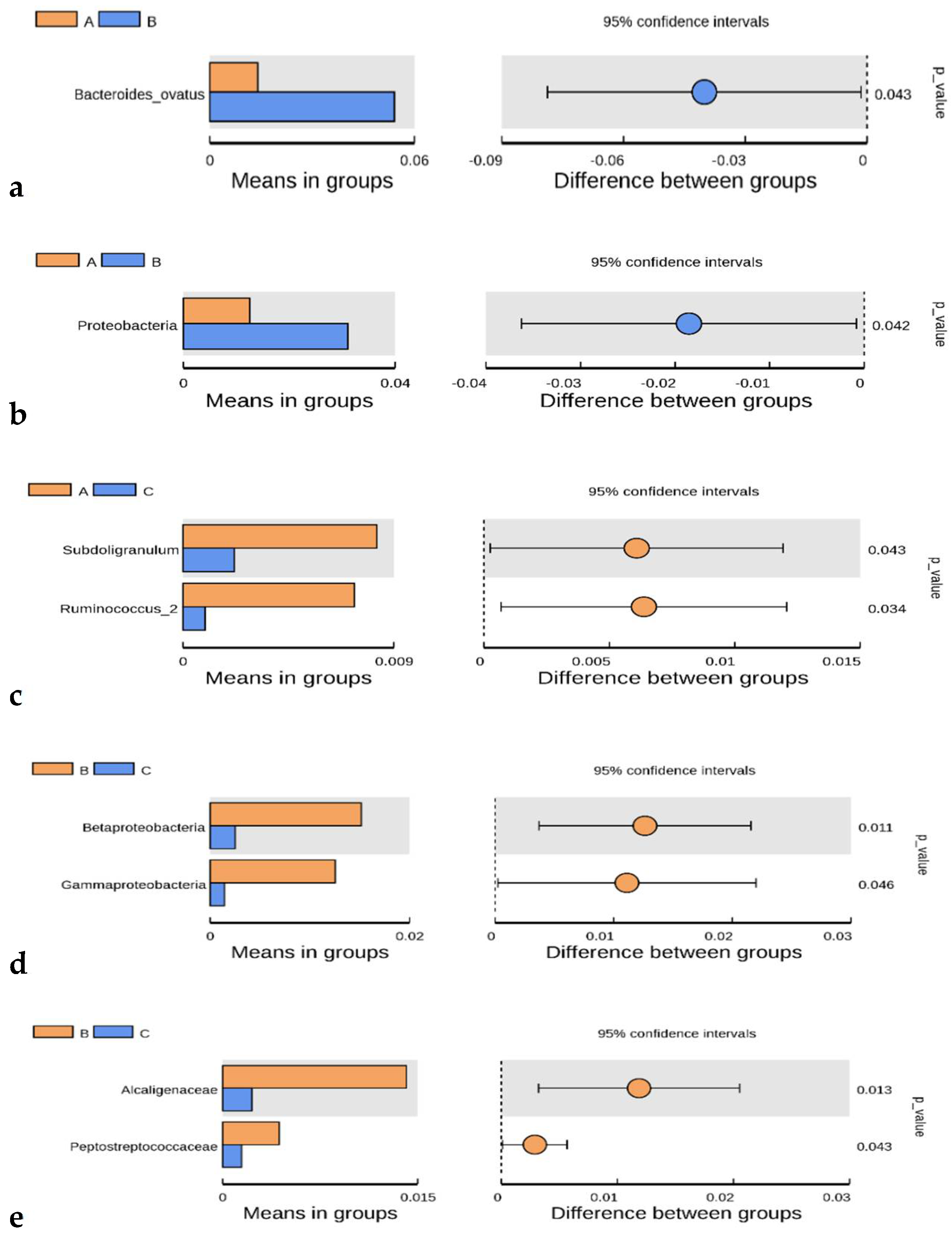
4.4. Differential Taxa and Biomarkers
4.5. LEfSe Biomarker Analysis
4.6. Future Directions
5. Conclusions
- The gut microbiota of patients with ASD, FXS, and FXS + ASD exhibits significant differences compared to that of neurotypical control individuals within the studied population.
- The microbiota of ASD patients is associated with an increased proportion of Firmicutes phylum members compared to the control group, a characteristic that, according to some authors, is linked to alterations in intestinal permeability.
- An increased abundance of the class Clostridia was observed in all three patient groups compared to controls, with this increase being most pronounced in FXS patients.
- Specific biomarkers were identified, distinguishing gut microbiota profiles of the patient groups from one another and from the control group.
- There are notable similarities in bacterial composition and abundance between the ASD and FXS + ASD groups, particularly regarding the species Bacteroides ovatus, which may be related to shared symptomatology.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD | Autism spectrum disorder |
| FXS | Fragile X Syndrome |
| DNA | Deoxyribonucleic acid |
| RRNA | Ribosomal ribonucleic acid |
| MetaHIT | Human Intestinal Tract Metagenomics |
| HMP | Human Microbiome Project |
| PCR | Polymerase Chain Reaction |
| CNS | Central nervous system |
| ANS | Autonomous Nervous System |
| HPA | Hypothalamic–Pituitary–Adrenal Axis |
| ENS | Enteric nervous system |
| OTU | Operative Taxonomic Unit |
| FLASH | Quick adjustment of the length of short reads to improve genome assemblies |
| ACP | Principal Component Analysis |
| ANOSIM | Analysis of Similarities |
| LDA | Linear Discriminant Analysis |
| LEfSe | Linear Discriminant Analysis Effect Size |
References
- Salcedo-Arellano, M.J.; Cabal-Herrera, A.M.; Punatar, R.H.; Clark, C.J.; Romney, C.A.; Hagerman, R.J. Overlapping molecular pathways leading to autism spectrum disorders, Fragile X syndrome, and targeted treatments. Neurotherapeutics 2021, 18, 265–283. [Google Scholar] [CrossRef]
- Riley, C.; Wheeler, A. Assessing the Fragile X Syndrome Newborn Screening Landscape. Pediatrics 2017, 139 (Suppl. S3), S207–S215. [Google Scholar] [CrossRef]
- Harris, S.W.; Hessl, D.; Goodlin-Jones, B.; Ferranti, J.; Bacalman, S.; Barbato, I.; Tassone, F.; Hagerman, P.J.; Herman, K.; Hagerman, R.J. Autism profiles of males with fragile X syndrome. Am. J. Ment. Retard. 2008, 113, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Kraan, C.M.; Godler, D.E.; Amor, D.J. Epigenetics of fragile X syndrome and fragile X-related disorders. Dev. Med. Child Neurol. 2019, 61, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Ribate Molina, M.P.; Pié Juste, J.; Ramos Fuentes, F.J. Síndrome de X Frágil. Protoc. Diagn. Ter. Pediatr. 2010, 1, 85–90. [Google Scholar]
- Kim, K.; Hessl, D.; Randol, J.L.; Espinal, G.M.; Schneider, A.; Protic, D.; Aydin, E.Y.; Hagerman, R.J.; Hagerman, P.J. Association between IQ and FMR1 protein (FMRP) across the spectrum of CGG repeat expansions. PLoS ONE 2019, 14, e0226811. [Google Scholar] [CrossRef]
- Suhl, J.A.; Chopra, P.; Anderson, B.R.; Bassell, G.J.; Warren, S.T. Analysis of FMRP mRNA target datasets reveals highly associated mRNAs mediated by G-quadruplex structures formed via clustered WGGA sequences. Hum. Mol. Genet. 2014, 23, 5479–5491. [Google Scholar] [CrossRef]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.S.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, S.; Delhaye, S.; Folci, A.; Paquet, A.; Brau, F.; Duprat, F.; Jarjat, M.; Grossi, M.; Béal, M.; Martin, S.; et al. New insights into the role of Cav2 protein family in calcium flux deregulation in Fmr1-KO neurons. Front. Mol. Neurosci. 2018, 11, 342. [Google Scholar] [CrossRef]
- El Bekay, R.; Romero-Zerbo, Y.; Decara, J.; Sánchez-Salido, L.; Del Arco-Herrera, I.; Rodríguez-de Fonseca, F.; De Diego-Otero, Y. Enhanced markers of oxidative stress, altered antioxidants and NADPH-oxidase activation in brains from Fragile X Mental Retardation 1-deficient mice, a pathological model for Fragile X syndrome. Eur. J. Neurosci. 2007, 26, 3169–3180. [Google Scholar] [CrossRef]
- De Diego-Otero, Y.; El Bekay, R.; García-Guirado, F.; Sánchez-Salido, L.; Giráldez-Pérez, R.M. Apocynin, a selective NADPH oxidase (Nox2) inhibitor, ameliorates behavioural and learning deficits in the Fragile X syndrome mouse model. Biomedicines 2024, 12, 2887. [Google Scholar] [CrossRef]
- Kau, A.S.M.; Tierney, E.; Bukelis, I.; Stump, M.H.; Kates, W.R.; Trescher, W.H.; Kaufmann, W.E. Social behavior profile in young males with fragile X syndrome: Characteristics and specificity. Am. J. Med. Genet. A 2004, 126, 9–17. [Google Scholar] [CrossRef]
- Aishworiya, R.; Chi, M.-H.; Zafarullah, M.; Mendoza, G.; Ponzini, M.D.; Kim, K.; Biag, H.M.B.; Thurman, A.J.; Abbeduto, L.; Hessl, D.; et al. Intercorrelation of molecular biomarkers and clinical phenotype measures in Fragile X syndrome. Cells 2023, 12, 1920. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, S.H.; Folsom, T.D. The role of Fragile X Mental Retardation Protein in major mental disorders. Neuropharmacology 2011, 60, 1221–1226. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.S. Cytogenetic abnormalities and fragile-X syndrome in autism spectrum disorder. BMC Med. Genet. 2005, 6, 3. [Google Scholar] [CrossRef]
- Altimiras, F.; Garcia, J.A.; Palacios-García, I.; Hurley, M.J.; Deacon, R.; González, B.; Cogram, P. Altered gut microbiota in a Fragile X syndrome mouse model. Front. Neurosci. 2021, 15, 653120. [Google Scholar] [CrossRef] [PubMed]
- Salmerón, A.M.; Pérez-Fernández, C.; Abreu, A.C.; Fernández, S.; Tristán, A.I.; Ruiz-Sobremazas, D.; Cabré, M.; Guardia-Escote, L.; Fernández, I.; Sánchez-Santed, F. Exploring microbiota-gut-brain axis biomarkers linked to autism spectrum disorder in prenatally chlorpyrifos-exposed Fmr1 knock-out and wild-type male rats. Toxicology 2024, 506, 153871. [Google Scholar] [CrossRef]
- Fattorusso, A.; Di Genova, L.; Dell’Isola, G.B.; Mencaroni, E.; Esposito, S. Autism spectrum disorders and the gut microbiota. Nutrients 2019, 11, 521. [Google Scholar] [CrossRef]
- Srikantha, P.; Mohajeri, M.H. The possible role of the microbiota-gut-brain axis in autism spectrum disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef]
- Bougeard, C.; Picarel-Blanchot, F.; Schmid, R.; Campbell, R.; Buitelaar, J. Prevalence of autism spectrum disorder and co-morbidities in children and adolescents: A systematic literature review. Front. Psychiatry 2021, 12, 744709. [Google Scholar] [CrossRef]
- Shaw, K.A.; Williams, S.; Patrick, M.E.; Valencia-Prado, M.; Durkin, M.S.; Howerton, E.M.; Ladd-Acosta, C.M.; Pas, E.T.; Bakian, A.V.; Bartholomew, P.; et al. Prevalence and Early Identification of Autism Spectrum Disorder Among Children Aged 4 and 8 Years—Autism and Developmental Disabilities Monitoring Network, 16 Sites, United States, 2022. MMWR Surveill. Summ. 2025, 74, 1–22. [Google Scholar] [CrossRef]
- Theoharides, T.C.; Kavalioti, M.; Martinotti, R. Factors adversely influencing neurodevelopment. J. Biol. Regul. Homeost. Agents 2019, 33, 1663–1667. [Google Scholar] [CrossRef]
- Yang, Y.; Tian, J.; Yang, B. Targeting gut microbiome: A novel and potential therapy for autism. Life Sci. 2018, 194, 111–119. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Kang, D.-W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-term benefit of microbiota transfer therapy on autism symptoms and gut microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef] [PubMed]
- Naviaux, R.K. Metabolic features of the cell danger response. Mitochondrion 2014, 16, 7–17. [Google Scholar] [CrossRef]
- van De Sande, M.M.H.; van Buul, V.J.; Brouns, F.J.P.H. Autism and nutrition: The role of the gut–brain axis. Nutr. Res. Rev. 2014, 27, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Fond, G.; Chevalier, G.; Eberl, G.; Leboyer, M. The potential role of microbiota in major psychiatric disorders: Mechanisms, preclinical data, gastro-intestinal comorbidities and therapeutic options. Presse Med. 2016, 45, 7–19. [Google Scholar] [CrossRef]
- Groer, M.W.; Gregory, K.E.; Louis-Jacques, A.; Thibeau, S.; Walker, W.A. The very low birth weight infant microbiome and childhood health. Birth Defects Res. Part C Embryo Today Rev. 2015, 105, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Callewaert, C.; Marotz, C.; Hyde, E.R.; Debelius, J.W.; McDonald, D.; Sogin, M.L. The microbiome and human biology. Annu. Rev. Genom. Hum. Genet. 2017, 18, 65–86. [Google Scholar] [CrossRef]
- Adamczak, A.M.; Werblińska, A.; Jamka, M.; Walkowiak, J. Maternal-Foetal/Infant Interactions-Gut Microbiota and Immune Health. Biomedicines 2024, 12, 490. [Google Scholar] [CrossRef]
- Thum, C.; Cookson, A.L.; Otter, D.E.; McNabb, W.C.; Hodgkinson, A.J.; Dyer, J.; Roy, N.C. Can nutritional modulation of maternal intestinal microbiota influence the development of the infant gastrointestinal tract? J. Nutr. 2012, 142, 1921–1928. [Google Scholar] [CrossRef]
- Theis, K.R.; Romero, R.; Winters, A.D.; Greenberg, J.M.; Gomez-Lopez, N.; Alhousseini, A.; Bieda, J.; Maymon, E.; Pacora, P.; Fettweis, J.M.; et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am. J. Obstet. Gynecol. 2019, 220, 267.e1–267.e39. [Google Scholar] [CrossRef]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed]
- Pantazi, A.C.; Balasa, A.L.; Mihai, C.M.; Chisnoiu, T.; Lupu, V.V.; Kassim, M.A.K.; Mihai, L.; Frecus, C.E.; Chirila, S.I.; Lupu, A.; et al. Development of Gut Microbiota in the First 1000 Days after Birth and Potential Interventions. Nutrients 2023, 15, 3647. [Google Scholar] [CrossRef]
- Sharon, G.; Cruz, N.J.; Kang, D.-W.; Gandal, M.J.; Wang, B.; Kim, Y.-M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human gut microbiota from autism spectrum disorder promote behavioural symptoms in mice. Cell 2019, 177, 1600–1618.e17. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota–gut–brain axis. Lancet Neurol. 2020, 19, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Marzal, L.N.; Rojas-Velazquez, D.; Rigters, D.; Prince, N.; Garssen, J.; Kraneveld, A.D.; Perez-Pardo, P.; Lopez-Rincon, A. A robust microbiome signature for autism spectrum disorder across different studies using machine learning. Sci. Rep. 2024, 14, 814. [Google Scholar] [CrossRef]
- Robles Alonso, V.; Guarner, F. Linking the gut microbiota to human health. Br. J. Nutr. 2013, 109, S21–S26. [Google Scholar] [CrossRef]
- Foster, J.A.; McVey Neufeld, K.A. Gut–brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef]
- Roubalová, R.; Procházková, P.; Papežová, H.; Smitka, K.; Bilej, M.; Tlaskalová-Hogenová, H. Anorexia nervosa: Gut microbiota–immune–brain interactions. Clin. Nutr. 2019, 39, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.T.; Lai, J.B.; Du, Y.L.; Xu, Y.; Ruan, L.M.; Hu, S.H. Current understanding of gut microbiota in mood disorders: An update of human studies. Front. Genet. 2019, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Xing, C.; Long, W.; Wang, H.Y.; Liu, Q.; Wang, R.F. Impact of microbiota on central nervous system and neurological diseases: The gut-brain axis. J. Neuroinflamm. 2019, 16, 53. [Google Scholar] [CrossRef]
- Li, Y.; Hao, Y.; Fan, F.; Zhang, B. The role of microbiome in insomnia, circadian disturbance and depression. Front. Psychiatry 2018, 9, 669. [Google Scholar] [CrossRef]
- Möhle, L.; Mattei, D.; Heimesaat, M.M.; Bereswill, S.; Fischer, A.; Alutis, M.; French, T.; Hambardzumyan, D.; Matzinger, P.; Dunay, I.R.; et al. Ly6Chi monocytes provide a link between antibiotic-induced changes in gut microbiota and adult hippocampal neurogenesis. Cell Rep. 2016, 15, 1945–1956. [Google Scholar] [CrossRef]
- Hill, C.J.; Lynch, D.B.; Murphy, K.; Ulaszewska, M.; Jeffery, I.B.; O’Shea, C.A.; Watkins, C.; Dempsey, E.; Mattivi, F.; Tuohy, K.; et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 2017, 5, 4. [Google Scholar] [CrossRef]
- Loh, J.S.; Mak, W.Q.; Tan, L.K.S.; Ng, C.X.; Chan, H.H.; Yeow, S.H.; Foo, J.B.; Ong, Y.S.; How, C.W.; Khaw, K.Y. Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases. Signal Transduct. Target. Ther. 2024, 9, 37. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Chao, A.; Shen, T.J. Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 2003, 10, 429–443. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Chao, A.; Hwang, W.-H.; Chen, Y.-C.; Kuo, C.-Y. Estimating the number of shared species in two communities. Stat. Sin. 2000, 10, 227–246. [Google Scholar]
- Wilcoxon, F. Individual comparisons of grouped data by ranking methods. J. Econ. Entomol. 1946, 39, 269–270. [Google Scholar] [CrossRef]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef]
- Fagerland, M.W. T-tests, non-parametric tests, and large studies—A paradox of statistical practice? BMC Med. Res. Methodol. 2012, 12, 78. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- De Lucas Moreno, B.; González Soltero, R.; Bressa, C.; Bailén, M.; Larrosa, M. Lifestyle modulation of gut microbiota. Nutr. Hosp. 2019, 36, 35–39. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Hughes, H.K.; Rose, D.; Ashwood, P. The gut microbiota and dysbiosis in autism spectrum disorders. Curr. Neurol. Neurosci. Rep. 2018, 18, 81. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Narzisi, A.; Masi, G.; Grossi, E. Nutrition and Autism Spectrum Disorder: Between False Myths and Real Research-Based Opportunities. Nutrients 2021, 13, 2068. [Google Scholar] [CrossRef]
- Argou-Cardozo, I.; Zeidán-Chuliá, F. Clostridium bacteria and autism spectrum conditions: A systematic review and hypothetical contribution of environmental glyphosate levels. Med. Sci. 2018, 6, 29. [Google Scholar] [CrossRef]
- Gabriele, S.; Sacco, R.; Cerullo, S.; Neri, C.; Urbani, A.; Tripi, G.; Malvy, J.; Barthelemy, C.; Bonnet-Brihault, F.; Persico, A.M. Urinary p-cresol is elevated in young French children with autism spectrum disorder: A replication study. Biomarkers 2014, 19, 463–470. [Google Scholar] [CrossRef]
- Protic, D.D.; Aishworiya, R.; Salcedo-Arellano, M.J.; Tang, S.J.; Milisavljevic, J.; Mitrovic, F.; Hagerman, R.J.; Budimirovic, D.B. Fragile X syndrome: From molecular aspect to clinical treatment. Int. J. Mol. Sci. 2022, 23, 1935. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Barnhill, K.; Devlin, M.; Moreno, H.T.; Potts, A.; Richardson, W.; Schutte, C.; Hewitson, L. Brief report: Implementation of a specific carbohydrate diet for a child with autism spectrum disorder and Fragile X syndrome. J. Autism Dev. Disord. 2020, 50, 1800–1808. [Google Scholar] [CrossRef]
- Mallozzi, M.; Viswanathan, V.K.; Vedantam, G. Spore-forming bacilli and clostridia in human disease. Future Microbiol. 2010, 5, 1109–1123. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Valdovinos-Díaz, M.A. Intestinal microbiota in digestive disorders: Probiotics, prebiotics and symbiotics. Rev. Gastroenterol. México 2013, 78, 25–27. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef]
- Kashtanova, D.A.; Tkacheva, O.N.; Doudinskaya, E.N.; Strazhesko, I.D.; Kotovskaya, Y.V.; Popenko, A.S.; Tyakht, A.V.; Alexeev, D.G. Gut microbiota in patients with different metabolic statuses: Moscow study. Microorganisms 2018, 6, 98. [Google Scholar] [CrossRef] [PubMed]
- Wijdeveld, M.; Nieuwdorp, M.; Ijzerman, R.G. The interaction between microbiome and host central nervous system: The gut-brain axis as a potential new therapeutic target in the treatment of obesity and cardiometabolic disease. Expert Opin. Ther. Targets 2020, 24, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef] [PubMed]







| Code | Group | Age (Years) | Gender | Phenotype |
|---|---|---|---|---|
| BTSXF20C | C | 10 | Male | Control |
| BTC23 | C | 8 | Female | Control |
| BTSXF16C | C | 10 | Male | Control |
| BTC004I | C | 4 | Male | Control |
| BTC07 | C | 10 | Male | Control |
| BTC10 | C | 5 | Male | Control |
| BTC02I2 | C | 8 | Female | Control |
| BTSXF02C | C | 3 | Male | Control |
| BTF3G02Cont | C | 8 | Male | Control |
| BTSXF19 | A | 6 | Male | FXS |
| BTSXF17 | A | 6 | Male | FXS |
| BTSXF15 | A | 6 | Male | FXS |
| BTSXF13 | A | 18 | Male | FXS |
| BTSXF01 | A | 3 | Male | FXS |
| BTSXF12 | A | 6 | Female | FXS |
| BT023Int | B | 6 | Male | ASD |
| BT021Int | B | 6 | Male | ASD |
| BT047 | B | 3 | Male | ASD |
| BT027Int | B | 6 | Male | ASD |
| BTH02Int2 | B | 8 | Male | ASD |
| BT046 | B | 4 | Male | ASD |
| BT061 | B | 5 | Male | ASD |
| BT004I | B | 4 | Male | ASD |
| BT012I | B | 3 | Male | ASD |
| BTSXF18 | F | 8 | Male | FXS + ASD |
| TRA152INT2 | F | 18 | Male | FXS + ASD |
| BTSXF05I | F | 4 | Male | FXS + ASD |
| BTSXF04 | F | 3 | Male | FXS + ASD |
| BTSXF9 | F | 3 | Female | FXS + ASD |
| BTSXF10 | F | 5 | Male | FXS + ASD |
| BTSXF11 | F | 3 | Male | FXS + ASD |
| Group | OTU # | Shannon | Simpson | CHAO1 | Good’s Coverage (%) |
|---|---|---|---|---|---|
| Control | 418.89 ± 120.04 | 3.85 ± 0.88 | 0.81 ± 0.14 | 478.39 ± 140.33 | 99.99 ± 0.00 |
| FXS | 424.00 ± 88.76 | 4.12 ± 0.86 | 0.90 ± 0.06 * | 473.25 ± 106.41 | 99.99 ± 0.00 |
| ASD | 460.70 ± 172.37 | 4.98 ± 0.57 ** | 0.92 ± 0.03 ** | 507.22 ± 184.34 | 99.99 ± 0.00 |
| FXS + ASD | 468.71 ± 168.96 | 4.70 ± 1.29 * | 0.89 ± 0.11 ** | 524.28 ± 162.40 | 99.99 ± 0.00 |
| Groups | Taxon | p-Value |
|---|---|---|
| ASD * vs. FXS | Bacteroides ovatus (Species) | 0.043 |
| ASD * vs. FXS | Proteobacteria (Phylum) | 0.042 |
| FXS * vs. Control | Subdoligranulum (Genus) | 0.043 |
| FXS * vs. Control | Ruminococcus (Genus) | 0.034 |
| ASD * vs. Control | Betaproteobacteria (Class) | 0.011 |
| ASD * vs. Control | Gammaproteobacteria (Class) | 0.046 |
| ASD * vs. Control | Alcaligenaceae (Family) | 0.013 |
| ASD * vs. Control | Peptostreptococcaceae (Family) | 0.043 |
| ASD * vs. Control | Subdoligranulum (Genus) | 0.049 |
| ASD * vs. Control | Intestinibacter (Genus) | 0.038 |
| ASD * vs. Control | Burkholderiales (Order) | 0.011 |
| ASD * vs. Control | Proteobacteria (Phylum) | 0.020 |
| FXS * vs. FXS + ASD | Fusicatenibacter (Genus) | 0.010 |
| ASD * vs. FXS + ASD | Peptostreptococcaceae (Family) | 0.046 |
| ASD * vs. FXS + ASD | Fusicatenibacter (Genus) | 0.046 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diego-Otero, Y.d.; Bodoque-García, A.; Quintero-Navarro, C.; Calvo-Medina, R.; Salgado-Cacho, J.M. An Altered Gut Microbiota–Brain Axis in Fragile X Syndrome May Explain Autistic Traits in Some Patients. Psychiatry Int. 2025, 6, 107. https://doi.org/10.3390/psychiatryint6030107
Diego-Otero Yd, Bodoque-García A, Quintero-Navarro C, Calvo-Medina R, Salgado-Cacho JM. An Altered Gut Microbiota–Brain Axis in Fragile X Syndrome May Explain Autistic Traits in Some Patients. Psychiatry International. 2025; 6(3):107. https://doi.org/10.3390/psychiatryint6030107
Chicago/Turabian StyleDiego-Otero, Yolanda de, Ana Bodoque-García, Carolina Quintero-Navarro, Rocío Calvo-Medina, and José María Salgado-Cacho. 2025. "An Altered Gut Microbiota–Brain Axis in Fragile X Syndrome May Explain Autistic Traits in Some Patients" Psychiatry International 6, no. 3: 107. https://doi.org/10.3390/psychiatryint6030107
APA StyleDiego-Otero, Y. d., Bodoque-García, A., Quintero-Navarro, C., Calvo-Medina, R., & Salgado-Cacho, J. M. (2025). An Altered Gut Microbiota–Brain Axis in Fragile X Syndrome May Explain Autistic Traits in Some Patients. Psychiatry International, 6(3), 107. https://doi.org/10.3390/psychiatryint6030107







