Nomogram Based on the Most Relevant Clinical, CT, and Radiomic Features, and a Machine Learning Model to Predict EGFR Mutation Status in Non-Small Cell Lung Cancer
Abstract
1. Introduction
- These procedures are often invasive;
- They may require repeated sampling due to the dynamic nature of tumor genetic mutations during treatment;
- Elderly patients and those with high-risk ground-glass opacity patterns often decline surgery or biopsy;
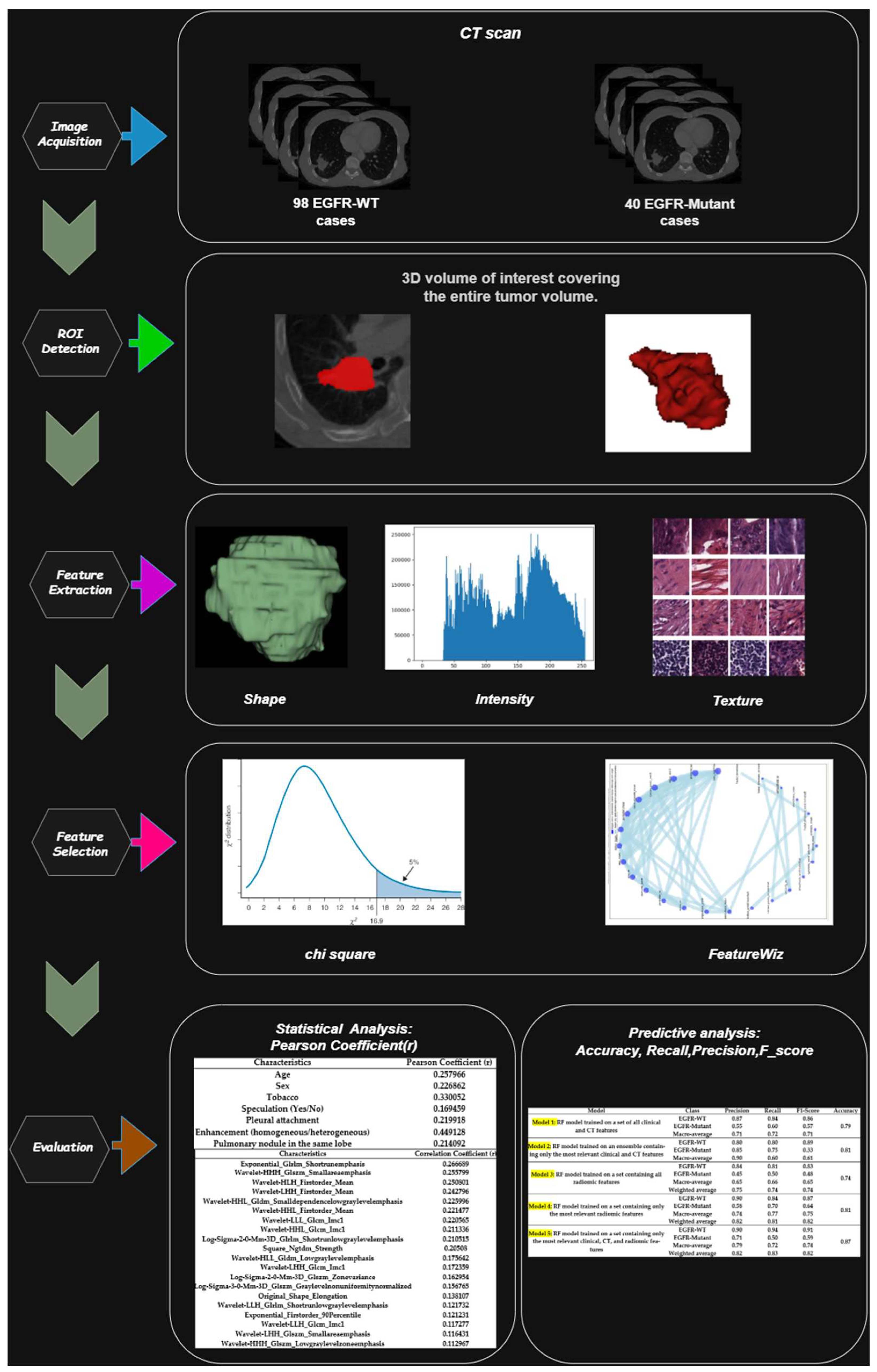
2. Materials and Methods
2.1. Study Design
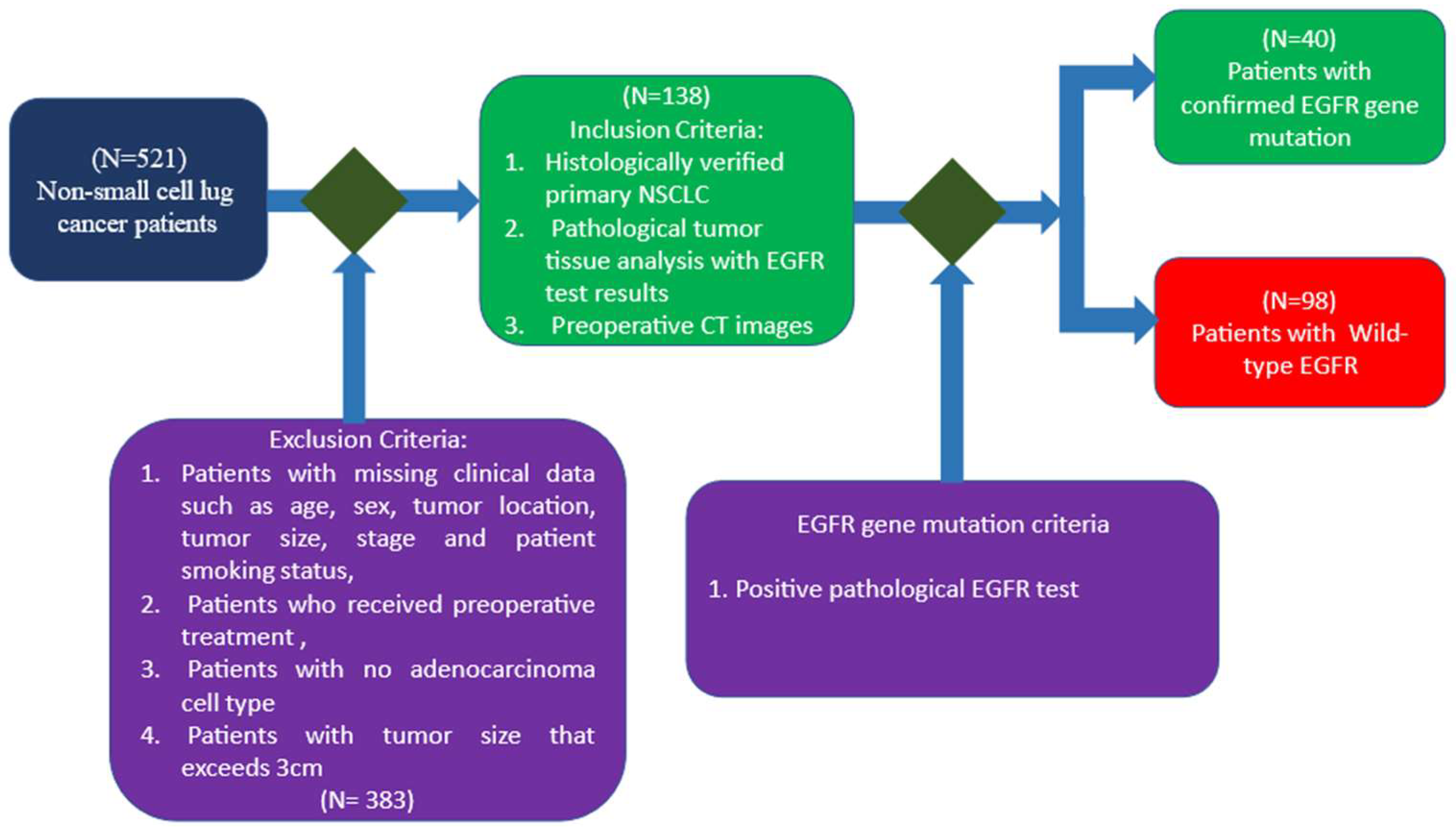
2.2. Patient Cohort and Data Collection
2.2.1. Inclusion and Exclusion Criteria
2.2.2. EGFR Mutation Evaluation Methods
2.3. Acquisition, Processing, and Segmentation of the CT Images
2.3.1. CT Image Acquisition
2.3.2. CT Image Processing
2.3.3. ROI Segmentation
2.4. Clinical, CT, and Radiomic Features
2.4.1. Clinical Characteristics
2.4.2. CT Characteristics
2.4.3. Radiomic Characteristics
2.4.4. Radiomic Feature Extraction
2.5. Development and Interpretability of Machine Learning Models
3. Results
3.1. Training and Testing Dataset
3.2. Statistical Analysis
3.3. Selection of the Most Relevant Clinical, CT, and Radiomic Features
3.4. Selection of Relevant Radiomic Characteristics
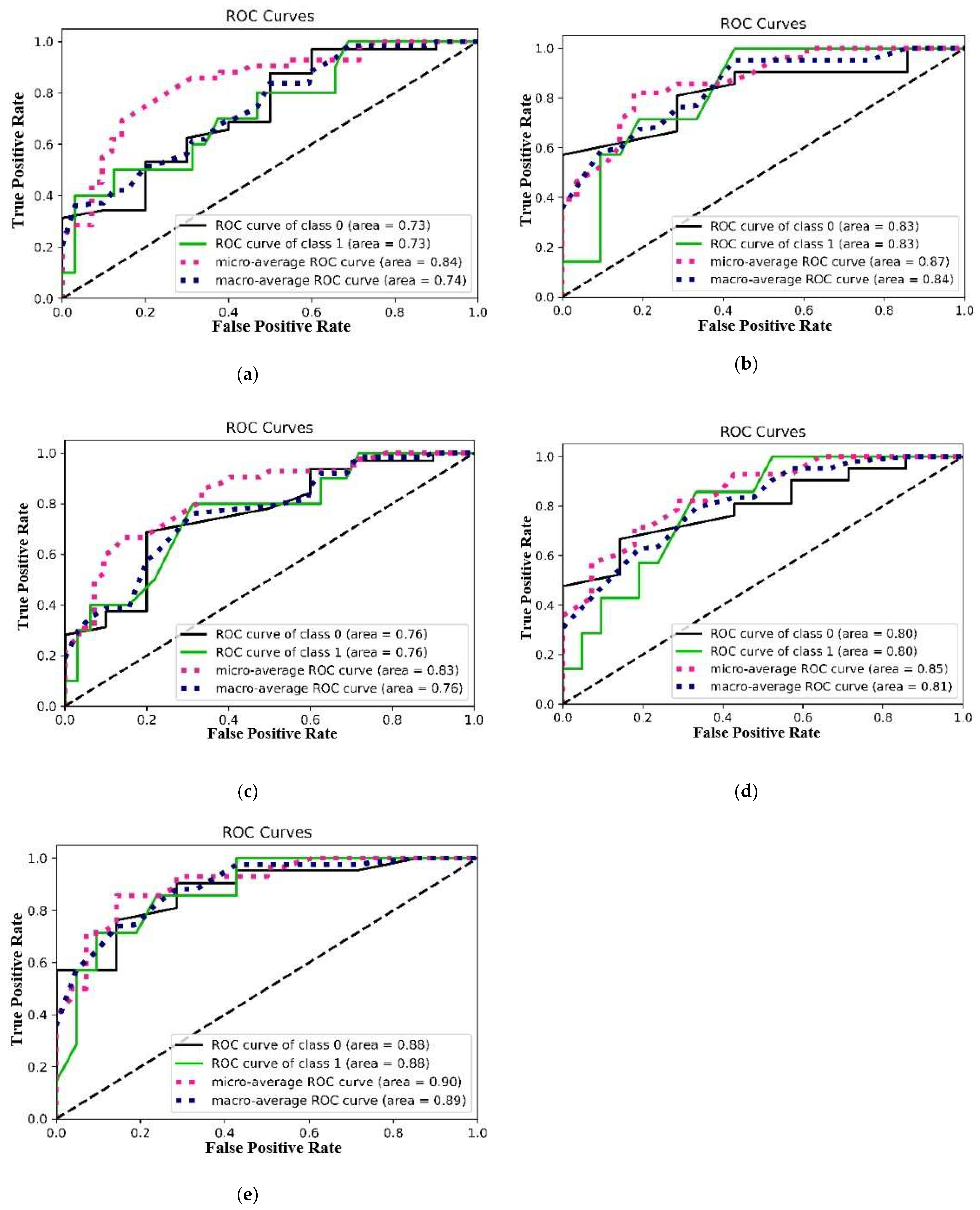
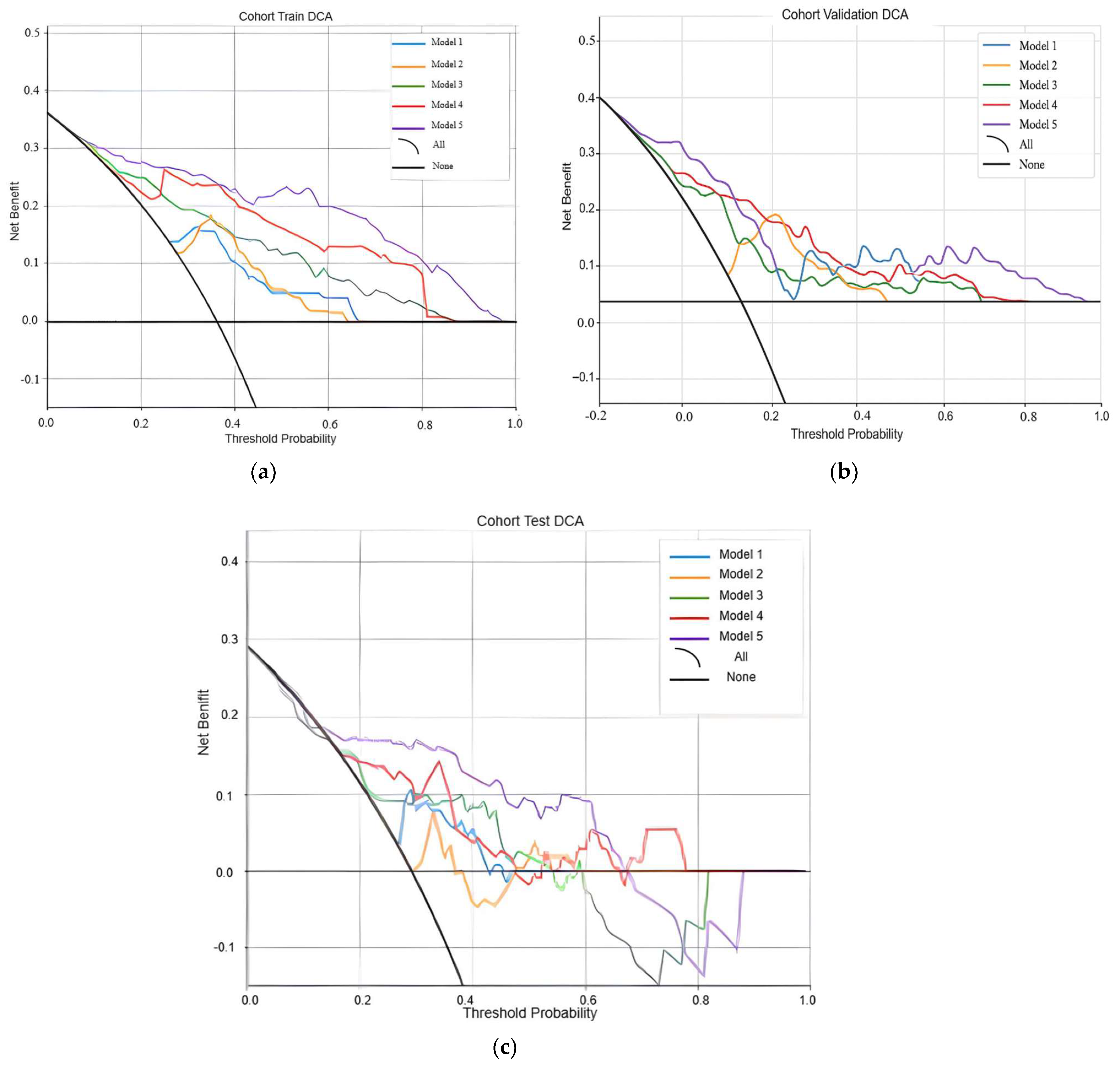
3.5. Measured Performances in the Testing Process
3.5.1. AUC/ROC Curve, Micro-Average, and Macro-Average ROC Curves
3.5.2. Decision Curves
3.5.3. DeLong Test
3.6. Interpretability
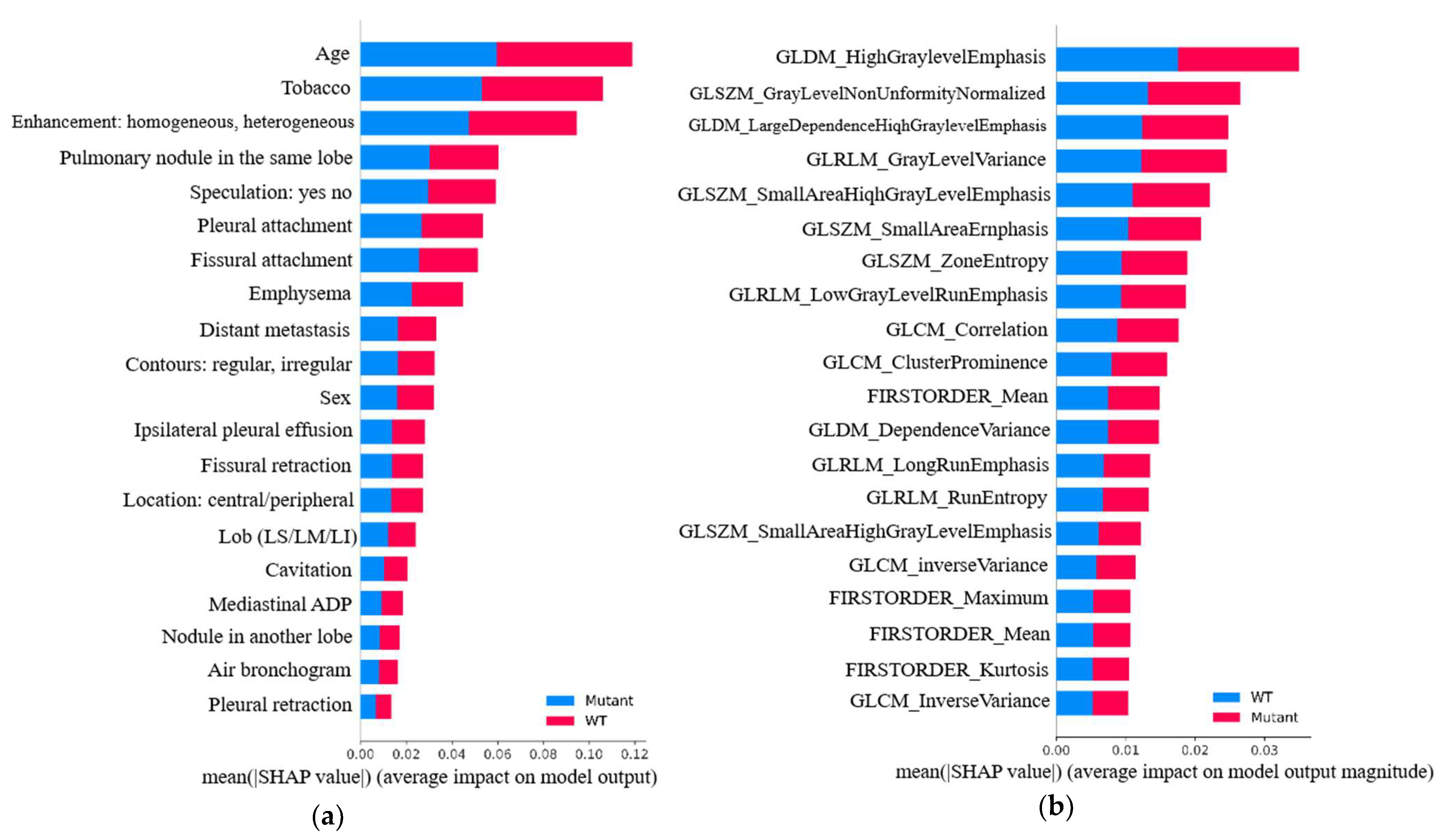
3.6.1. Global Relevance
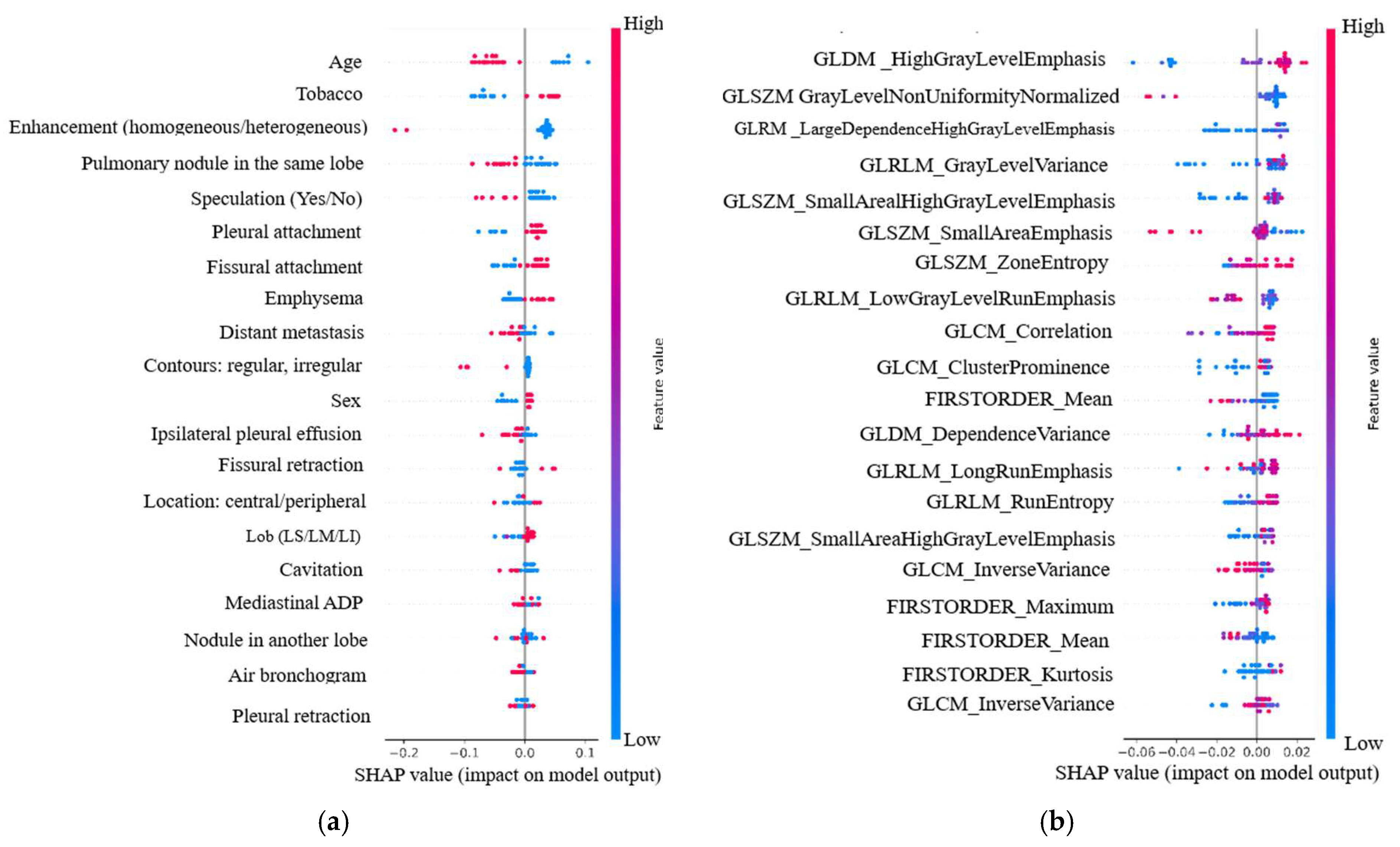
3.6.2. Individual Relevance
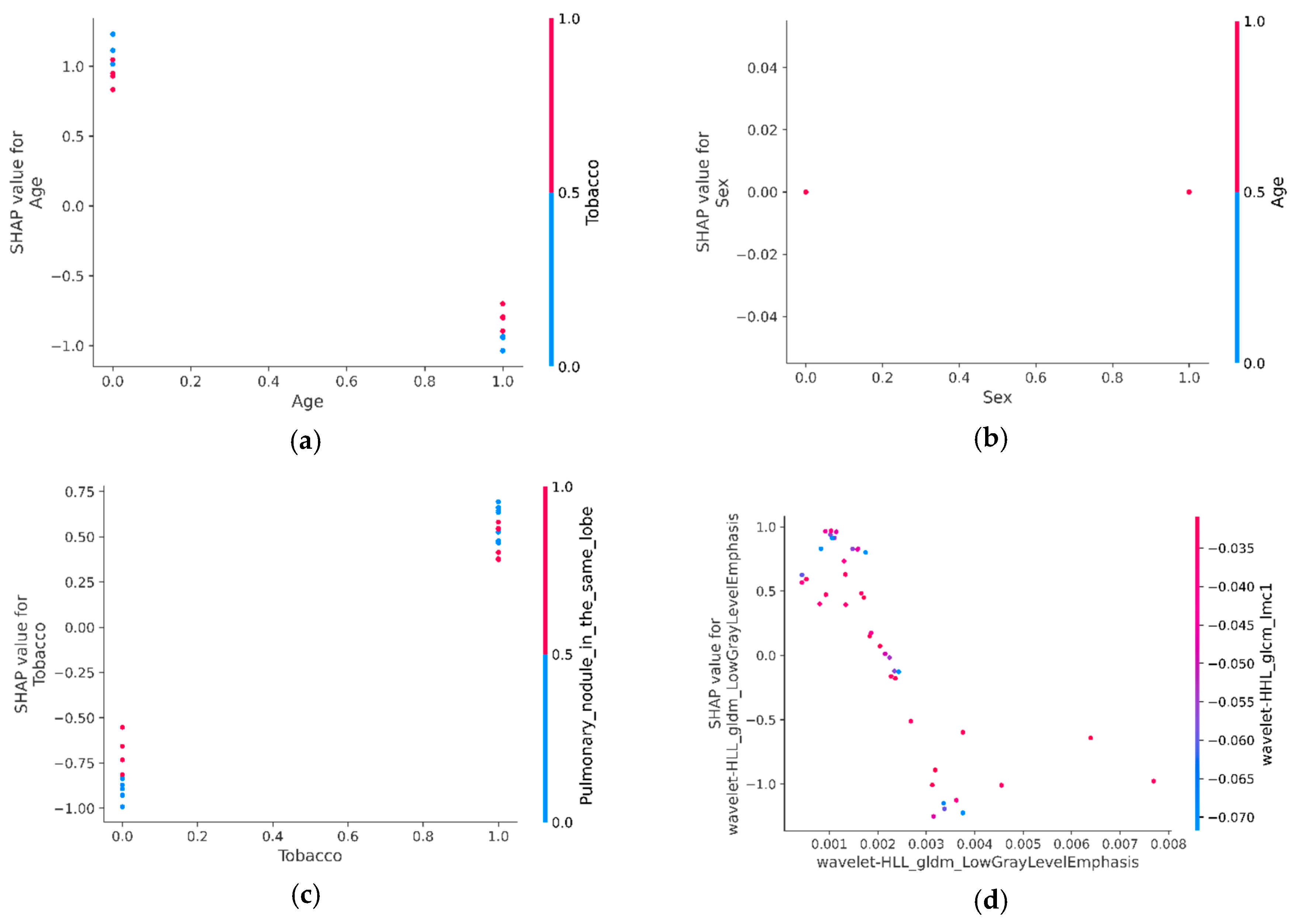
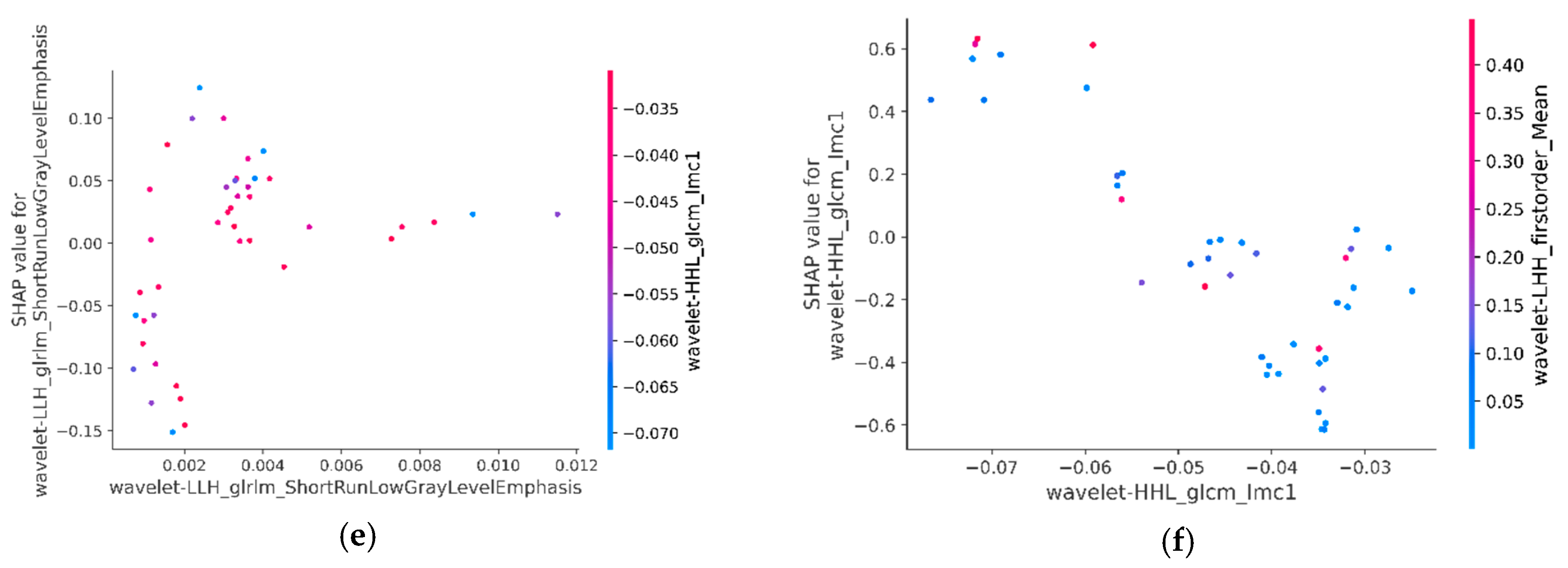
3.6.3. SHAP Interaction Values of Most Relevant Characteristics with Each Other
4. Discussion
4.1. Key Findings and Implications
4.2. Challenges and Future Directions
4.3. Significance and Originality
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EGFR | Epithelial Growth Factor Receptor |
| TKI | Tyrosine Kinase Inhibitor |
| NSCLC | Non-small cell lung cancer |
| AUC | Area under the curve |
| WT | Wild type |
| DICOM | Digital imaging and communications in medicine |
| SHAP | SHapley Additive exPlanations algorithm |
| ROC | Receiver Operating Characteristic |
| CT | Computed Tomography |
References
- Santos, D.C.; Saieg, M.A.; Geddie, W.; Leighl, N. EGFR gene status in cytological samples of non small cell lung carcinoma: Controversies and opportunities. Cancer Cytopathol. 2011, 119, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Ladanyi, M.; Pao, W. Lung adenocarcinoma: Guiding EGFR-targeted therapy and beyond. Mod. Pathol. 2008, 21 (Suppl. 2), S16–S22. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The Biology and Management of Non-Small Cell Lung Cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sheng, Z.; Zhang, J.; Song, J.; Teng, L.; Liu, L.; Li, Q.; Wang, B.; Li, B. Comparison of lorlatinib, alectinib and brigatinib in ALK inhibitor–naive/untreated ALK-positive advanced non-small-cell lung cancer: A systematic review and network meta-analysis. J. Chemother. 2021, 34, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; van Zandwijk, N.; Gridelli, C.; Baliko, Z.; Rischin, D.; Allan, S.; Krzakowski, M.; Heigener, D. Erlotinib in advanced non-small cell lung cancer: Efficacy and safety findings of the global phase IV Tarceva Lung Cancer Survival Treatment study. J. Thorac. Oncol. 2010, 5, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Cullen, M.H.; Zatloukal, P.; Sörenson, S.; Novello, S.; Fischer, J.R.; Joy, A.A.; Zereu, M.; Peterson, P.; Visseren-Grul, C.M.; Iscoe, N. A randomized phase III trial comparing standard and high-dose pemetrexed as second-line treatment in patients with locally advanced or metastatic non-small-cell lung cancer. Ann. Oncol. 2008, 19, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, M.; Yano, S.; Giaccone, G.; Tamura, T.; Nakagawa, K.; Douillard, J.Y.; Nishiwaki, Y.; Vansteenkiste, J.; Kudoh, S.; Rischin, D. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 trial) corrected. J. Clin. Oncol. 2003, 21, 2237–2246. [Google Scholar] [CrossRef] [PubMed]
- Kris, M.G.; Natale, R.B.; Herbst, R.S.; Lynch, T.J., Jr.; Prager, D.; Belani, C.P.; Schiller, J.H.; Kelly, K.; Spiridonidis, H.; Sandler, A. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: A randomized trial. JAMA 2003, 290, 2149–2158. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Soler, R.; Chachoua, A.; Hammond, L.A.; Rowinsky, E.K.; Huberman, M.; Karp, D.; Rigas, J.; Clark, G.M.; Santabárbara, P.; Bonomi, P. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J. Clin. Oncol. 2004, 22, 3238–3247. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.A.; Rodrigues Pereira, J.; Ciuleanu, T.E.; Tan, E.H.; Hirsh, V.; Thongprasert, S.; Campos, D.; Maoleekoonpiroj, S.; Smylie, M.; Martins, R.; et al. Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 2005, 353, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Riely, G.J.; Pao, W.; Pham, D.; Li, A.R.; Rizvi, N.; Venkatraman, E.S.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M.; Miller, V.A. Clinical Course of Patients with Non–Small Cell Lung Cancer and Epidermal Growth Factor Receptor Exon 19 and Exon 21 Mutations Treated with Geftinib or Erlotinib. Cancer Ther. Clin. 2006, 12, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.; Wu, Y.-L.; Hirsh, V.; O’bYrne, K.; Yamamoto, N.; Mok, T.; Popat, S.; Sequist, L.V.; Massey, D.; Zazulina, V.; et al. First-line afatinib versus chemotherapy in patients with non-small cell lung cancer and common epidermal growth factor receptor gene mutations and brain metastases. J. Thorac. Oncol. 2016, 11, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Rueschhoff, A.B.; Moore, A.W.; Jasahui, M.R.P. Lung Cancer Staging—A Clinical Practice Review. J. Respir. 2024, 4, 50–61. [Google Scholar] [CrossRef]
- Tang, W.; Li, X.; Xie, X.; Sun, X.; Liu, J.; Zhang, J.; Wang, C.; Yu, J.; Xie, P. EGFR Inhibitors as Adjuvant Therapy for Resected Non-Small Cell Lung Cancer Harboring EGFR Mutations. Lung Cancer 2019, 136, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Mitsudomi, T.; Morita, S.; Yatabe, Y.; Negoro, S.; Okamoto, I.; Tsurutani, J.; Seto, T.; Satouchi, M.; Tada, H.; Hirashima, T.; et al. Gefitinib Versus Cisplatin Plus Docetaxel in Patients with Non-Small-Cell Lung Cancer Harbouring Mutations of the Epidermal Growth Factor Receptor (Wjtog3405): An Open Label, Randomised Phase 3 Trial. Lancet Oncol. 2010, 11, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Herrera, J.; Montero-Fernandez, M.A.; Kokaraki, G.; De Petris, L.; Falcão, R.M.; Molina-Centelles, M.; Guijarro, R.; Ekman, S.; Ortiz-Villalón, C. NTRK Gene Expression in Non-Small-Cell Lung Cancer. J. Respir. 2025, 5, 2. [Google Scholar] [CrossRef]
- Kobayashi, K. Primary Resistance to EGFR Tyrosine Kinase Inhibitors (TKIs): Contexts and Comparisons in EGFR-Mutated Lung Cancer. J. Respir. 2023, 3, 223–236. [Google Scholar] [CrossRef]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non–small cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Brody, R.; Zhang, Y.; Ballas, M.; Siddiqui, M.K.; Gupta, P.; Barker, C.; Midha, A.; Walker, J. PD-L1 Expression in Advanced NSCLC: Insights into Risk Stratification and Treatment Selection from a Systematic Literature Review. Lung Cancer 2017, 112, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Pallumeera, M.; Giang, J.C.; Singh, R.; Pracha, N.S.; Makary, M.S. Evolving and Novel Applications of Artificial Intelligence in Cancer Imaging. Cancers 2025, 17, 1510. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Salehjahromi, M.; Godoy, M.C.B.; Qin, K.; Plummer, C.M.; Zhang, Z.; Hong, L.; Heeke, S.; Le, X.; Vokes, N.; et al. Lung Cancer Risk Prediction in Patients with Persistent Pulmonary Nodules Using the Brock Model and Sybil Model. Cancers 2025, 17, 1499. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, K.; Kawauchi, S.; Okamoto, M.; Tanabe, K.; Hayashi, C.; Mikami, M.; Kusumoto, T. Comprehensive Serum Glycopeptide Spectra Analysis Combined with Machine Learning for Early Detection of Lung Cancer: A Case–Control Study. Cancers 2025, 17, 1474. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, N.; Carrillo-Perez, F.; Ortuño, F.M.; Rojas, I.; Valenzuela, O. Understanding the Impact of Deep Learning Model Parameters on Breast Cancer Histopathological Classification Using ANOVA. Cancers 2025, 17, 1425. [Google Scholar] [CrossRef] [PubMed]
- Şeker, M.; Niazi, M.K.K.; Chen, W.; Frankel, W.L.; Gurcan, M.N. Tumor Bud Classification in Colorectal Cancer Using Attention-Based Deep Multiple Instance Learning and Domain-Specific Foundation Models. Cancers 2025, 17, 1245. [Google Scholar] [CrossRef] [PubMed]
- Marquette, C.-H.; Boutros, J.; Benzaquen, J.; Ferreira, M.; Pastre, J.; Pison, C.; Padovani, B.; Bettayeb, F.; Fallet, V.; Guibert, N.; et al. Circulating tumor cells as a potential biomarker for lung cancer screening: A prospective cohort study. Lancet Respir. Med. 2020, 8, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Duranti, L.; Tavecchio, L.; Rolli, L.; Solli, P. New Perspectives on Lung Cancer Screening and Artificial Intelligence. Life 2025, 15, 498. [Google Scholar] [CrossRef] [PubMed]
- Benfares, A.; Mourabiti, A.Y.; Alami, B.; Boukansa, S.; El Bouardi, N.; Lamrani, M.Y.A.; El Fatimi, H.; Amara, B.; Serraj, M.; Mohammed, S.; et al. Non-invasive, fast, and high-performance EGFR gene mutation prediction method based on deep transfer learning and model stacking for patients with Non-Small Cell Lung Cancer. Eur. J. Radiol. Open 2024, 13, 100601, ISSN 2352-0477. [Google Scholar] [CrossRef] [PubMed]
- Sait, W.A.R. Lung Cancer Detection Model Using Deep Learning Technique. Appl. Sci. 2023, 13, 12510. [Google Scholar] [CrossRef]
- Wang, L. Deep Learning Techniques to Diagnose Lung Cancer. Cancers 2022, 14, 5569. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Li, Y.; Xu, X.; Zheng, Z.; Li, F.; Fang, K.; Wang, Y.; Wang, B.; Hou, D. Multi sequence MRI-based radiomics nomogram for early prediction of osimertinib resistance in patients with non-small cell lung cancer brain metastases’, European. J. Radiol. Open 2023, 11, 100521. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, A.F.; Camacho, T.C.; Vásquez, A.F.; del Castillo Herazo, V.; Neira, J.G.A.; Yepes, M.M.; Camacho, E.C. Clinical and CT patterns to predict EGFR mutation in patients with non-small cell lung cancer: A systematic literature review and meta-analysis. Eur. J. Radiol. Open 2022, 9, 100400. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Xiong, X.; Wang, H.; Li, W. Explainable Machine Learning Model to Prediction EGFR Mutation. Lung Cancer 2022, 12, 924144. [Google Scholar] [CrossRef]
- Yang, C.; Chen, W.; Gong, G.; Li, Z.; Qiu, Q.; Yin, Y. Application of CT radiomics features to predict the EGFR mutation status and therapeutic sensitivity to TKIs of advanced lung adenocarcinoma. Transl. Cancer Res. 2020, 9, 6683–6690. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhang, G.; Lin, J.; Pang, Y.; Wang, H.; Bai, T.; Zhong, L. Multi-modal feature-fusion for CT metal artifact reduction using edge-enhanced generative adversarial networks. Comput. Methods Programs Biomed. 2022, 217, 106700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-K.; Zhang, Y.-G.; Zhou, Z.; Li, Y.-F. HONGAT: Graph Attention Networks in the Presence of High-Order Neighbors. In Proceedings of the AAAI Conference on Artificial Intelligence, Philadelphia, PA, USA, 25 February–4 March 2025; Volume 38, pp. 16750–16758. [Google Scholar] [CrossRef]
- Roy, S.; Carass, A.; Bazin, P.-L.; Prince, J.L.; Dawant, B.M.; Haynor, D.R. Intensity Inhomogeneity Correction of Magnetic Resonance Images using Patches. Proc. SPIE Int. Soc. Opt. Eng. 2011, 7962, 444–449. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yushkevich, P.A.; Gao, Y.; Gerig, G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; Volume 2016, pp. 3342–3345. [Google Scholar] [CrossRef]
- Egger, J.; Kapur, T.; Fedorov, A.; Pieper, S.; Miller, J.V.; Veeraraghavan, H.; Freisleben, B.; Golby, A.J.; Nimsky, C.; Kikinis, R. GBM volumetry using the 3D slicer-medical image computing platform. Sci. Rep. 2013, 3, 1364. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, E.R.; Parmar, C.; Jermoumi, M.; Mak, R.H.; van Baardwijk, A.; Fennessy, F.M.; Lewis, J.H.; De Ruysscher, D.; Kikinis, R.; Lambin, P.; et al. Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Sci. Rep. 2013, 3, 3529. [Google Scholar] [CrossRef] [PubMed]
- MacMahon, H.; Naidich, D.P.; Goo, J.M.; Lee, K.S.; Leung, A.N.C.; Mayo, J.R.; Mehta, A.C.; Ohno, Y.; Powell, C.A.; Prokop, M.; et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society. Radioloy 2017, 284, 228–243. [Google Scholar]
- Rubin, G.D. Lung Nodule and Cancer Detection in Computed Tomography Screening. J. Thorac. Imaging 2015, 30, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Benfante, V.; Salvaggio, G.; Ali, M.; Cutaia, G.; Salvaggio, L.; Salerno, S.; Busè, G.; Tulone, G.; Pavan, N.; Di Raimondo, D.; et al. Grading and Staging of Bladder Tumors Using Radiomics Analysis in Magnetic Resonance Imaging. In Image Analysis and Processing—ICIAP 2023 Workshops; Foresti, G.L., Fusiello, A., Hancock, E., Eds.; ICIAP 2023, Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2024; Volume 14366. [Google Scholar] [CrossRef]
- Canfora, I.; Cutaia, G.; Marcianò, M.; Calamia, M.; Faraone, R.; Cannella, R.; Benfante, V.; Comelli, A.; Guercio, G.; Giuseppe, L.R.; et al. A Predictive System to Classify Preoperative Grading of Rectal Cancer Using Radiomics Features. In Image Analysis and Processing; Mazzeo, P.L., Frontoni, E., Sclaroff, S., Distante, C., Eds.; ICIAP 2022 Workshops, ICIAP 2022, Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2022; Volume 13373. [Google Scholar] [CrossRef]
- Kiso, T.; Okada, Y.; Kawata, S.; Shichiji, K.; Okumura, E.; Hatsumi, N.; Matsuura, R.; Kaminaga, M.; Kuwano, H.; Okumura, E. Ultrasound-based radiomics and machine learning for enhanced diagnosis of knee osteoarthritis: Evaluation of diagnostic accuracy, sensitivity, specificity, and predictive value. Eur. J. Radiol. Open 2025, 14, 100649, ISSN 2352-0477. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Huang, Q.; Zhu, J.; Huang, W.; Wu, Q.; Fu, T.; Peng, S.; Zou, J. Prediction of plaque progression using different machine learning models of pericoronary adipose tissue radiomics based on coronary computed tomography angiography. Eur. J. Radiol. Open 2025, 14, 100638, ISSN 2352-0477. [Google Scholar] [CrossRef] [PubMed]
- Elizabeth, P.V.; Wong, M.Y.Z.; Leonardo, R.; Tarkin, J.M.; Evans, N.R.; Weir-McCall, J.R.; Chowdhury, M.M.; Coughlin, P.A.; Pavey, H.; Zaccagna, F.; et al. Using machine learning to predict carotid artery symptoms from CT angiography: A radiomics and deep learning approach. Eur. J. Radiol. Open 2024, 13, 100594, ISSN 2352-0477. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Rodrigues, C.F.d.S.; de Lima, F.J.C.; Barbosa, F.T. Importance of using basic statistics adequately in clinical research. J. Anesthesiol. 2017, 67, 619–625. [Google Scholar] [CrossRef]
- Seshadri, R. Github—Autoviml/Featurewiz: Use Advanced Feature Engineering Strategies And Select The Best Features From Your Data Set Fast with a Single Line of Code. 2020. Available online: https://github.com/AutoViML/featurewiz (accessed on 28 March 2025).
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 30th Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–8 December 2017. [Google Scholar]
- Van Sanden, S.; Murton, M.; Bobrowska, A.; Rahhali, N.; Sermon, J.; Rodrigues, B.; Goff-Leggett, D.; Chouaid, C.; Sebastian, M.; Greystoke, A. Greystoke. Prevalence of Epidermal Growth Factor Receptor Exon 20 Insertion Mutations in Non-small-Cell Lung Cancer in Europe: A Pragmatic Literature Review and Meta-analysis. Target Oncol. 2022, 17, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Sow, M.L.; El Yacoubi, H.; Moukafih, B.; Balde, S.; Akimana, G.; Najem, S.; El Khoyaali, S.; Abahssain, H.; Chaibi, A.; Khan, S.Z.; et al. Frequency and types of EGFR mutations in Moroccan patients with non–small cell lung cancer. Tumori 2021, 107, 335–340. [Google Scholar] [CrossRef]
- Deng, C.; Zhang, Y.; Ma, Z.; Fu, F.; Deng, L.; Li, Y.; Chen, H. Prognostic value of epidermal growth factor receptor gene mutation in resected lung adenocarcinoma. J. Thorac. Cardiovasc. Surg. 2021, 162, 664–674.e7. [Google Scholar] [CrossRef] [PubMed]
- Aokage, K.; Miyoshi, T.; Wakabayashi, M.; Ikeno, T.; Suzuki, J.; Tane, K.; Samejima, J.; Tsuboi, M. Prognostic influence of epidermal growth factor receptor mutation and radiological ground glass appearance in patients with early-stage lung adenocarcinoma. Lung Cancer 2021, 160, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Kamigaichi, A.; Mimae, T.; Tsubokawa, N.; Miyata, Y.; Adachi, H.; Shimada, Y.; Ito, H.; Ikeda, N.; Okada, M. Risk Factors for Recurrence of Stage I Epidermal Growth Factor Receptor Mutated Lung Adenocarcinoma. Ann. Thorac. Surg. 2024, 117, 743–751. [Google Scholar] [CrossRef] [PubMed]
| Random Forest Model | Dataset Composition |
|---|---|
| Model 1 | All clinical and CT features |
| Model 2 | Most relevant clinical and CT selected features |
| Model 3 | All radiomic features |
| Model 4 | Most relevant radiomic features |
| Model 5 | Most relevant clinical, CT, and radiomic features |
| Characteristics | Pearson Coefficient (r) |
|---|---|
| Age | 0.257966 |
| Sex | 0.226862 |
| Tobacco | 0.330052 |
| Speculation (Yes/No) | 0.169459 |
| Pleural attachment | 0.219918 |
| Enhancement (homogeneous/heterogeneous) | 0.449128 |
| Pulmonary nodule in the same lobe | 0.214092 |
| Characteristics | p-Value | CI 95% |
|---|---|---|
| Age | 0.01 | [0.219, 0.730] |
| Sex | 0.0006 | [0.232, 0.778] |
| Tobacco | 0.00 | [0.344, 0.982] |
| Speculation (Yes/No) | 0.043 | [0.214, 0.608] |
| Pleural attachment | 0.009 | [0.222, 0.742] |
| Enhancement (homogeneous/heterogeneous) | 0.0 | [0.442, 0.999] |
| Pulmonary nodule in the same lobe | 0.021 | [0.197, 0.648] |
| Characteristics | Correlation Coefficient (r) |
|---|---|
| Exponential_Glrlm_Shortrunemphasis | 0.266689 |
| Wavelet-HHH_Glszm_Smallareaemphasis | 0.255799 |
| Wavelet-HLH_Firstorder_Mean | 0.250801 |
| Wavelet-LHH_Firstorder_Mean | 0.242796 |
| Wavelet-HHL_Gldm_Smalldependencelowgraylevelemphasis | 0.225996 |
| Wavelet-HHL_Firstorder_Mean | 0.221477 |
| Wavelet-LLL_Glcm_Imc1 | 0.220565 |
| Wavelet-HHL_Glcm_Imc1 | 0.211336 |
| Log-Sigma-2-0-Mm-3D_Glrlm_Shortrunlowgraylevelemphasis | 0.210515 |
| Square_Ngtdm_Strength | 0.20508 |
| Wavelet-HLL_Gldm_Lowgraylevelemphasis | 0.175642 |
| Wavelet-LHH_Glcm_Imc1 | 0.172359 |
| Log-Sigma-2-0-Mm-3D_Glszm_Zonevariance | 0.162954 |
| Log-Sigma-3-0-Mm-3D_Glszm_Graylevelnonuniformitynormalized | 0.156765 |
| Original_Shape_Elongation | 0.138107 |
| Wavelet-LLH_Glrlm_Shortrunlowgraylevelemphasis | 0.121732 |
| Exponential_Firstorder_90Percentile | 0.121231 |
| Wavelet-LLH_Glcm_Imc1 | 0.117277 |
| Wavelet-LHH_Glszm_Smallareaemphasis | 0.116431 |
| Wavelet-HHH_Glszm_Lowgraylevelzoneemphasis | 0.112967 |
| Model | Class | Precision | Recall | F1-Score | Accuracy |
|---|---|---|---|---|---|
| Model 1: RF model trained on a set of all clinical and CT features | EGFR-WT | 0.87 | 0.84 | 0.86 | 0.79 |
| EGFR-Mutant | 0.55 | 0.60 | 0.57 | ||
| Macro-average | 0.71 | 0.72 | 0.71 | ||
| Model 2: RF model trained on an ensemble containing only the most relevant clinical and CT features | EGFR-WT | 0.80 | 0.80 | 0.89 | 0.81 |
| EGFR-Mutant | 0.85 | 0.75 | 0.33 | ||
| Macro-average | 0.90 | 0.60 | 0.61 | ||
| Model 3: RF model trained on a set containing all radiomic features | EGFR-WT | 0.84 | 0.81 | 0.83 | 0.74 |
| EGFR-Mutant | 0.45 | 0.50 | 0.48 | ||
| Macro-average | 0.65 | 0.66 | 0.65 | ||
| Weighted average | 0.75 | 0.74 | 0.74 | ||
| Model 4: RF model trained on a set containing only the most relevant radiomic features | EGFR-WT | 0.90 | 0.84 | 0.87 | 0.81 |
| EGFR-Mutant | 0.58 | 0.70 | 0.64 | ||
| Macro-average | 0.74 | 0.77 | 0.75 | ||
| Weighted average | 0.82 | 0.81 | 0.82 | ||
| Model 5: RF model trained on a set containing only the most relevant clinical, CT, and radiomic features | EGFR-WT | 0.90 | 0.94 | 0.91 | 0.87 |
| EGFR-Mutant | 0.71 | 0.50 | 0.59 | ||
| Macro-average | 0.79 | 0.72 | 0.74 | ||
| Weighted average | 0.82 | 0.83 | 0.82 |
| Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | |
|---|---|---|---|---|---|
| Model 1 | 1 | 0.030 | 0.684 | 0.094 | 0.008 |
| Model 2 | 0.030 | 1 | 0.057 | 0.471 | 0.211 |
| Model 3 | 0.684 | 0.057 | 1 | 0.193 | 0.011 |
| Model 4 | 0.094 | 0.471 | 0.193 | 1 | 0.044 |
| Model 5 | 0.008 | 0.211 | 0.01 | 0.044 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benfares, A.; Mourabiti, A.y.; Alami, B.; Boukansa, S.; Benomar, I.; El Bouardi, N.; Alaoui Lamrani, M.Y.; El Fatimi, H.; Amara, B.; Serraj, M.; et al. Nomogram Based on the Most Relevant Clinical, CT, and Radiomic Features, and a Machine Learning Model to Predict EGFR Mutation Status in Non-Small Cell Lung Cancer. J. Respir. 2025, 5, 11. https://doi.org/10.3390/jor5030011
Benfares A, Mourabiti Ay, Alami B, Boukansa S, Benomar I, El Bouardi N, Alaoui Lamrani MY, El Fatimi H, Amara B, Serraj M, et al. Nomogram Based on the Most Relevant Clinical, CT, and Radiomic Features, and a Machine Learning Model to Predict EGFR Mutation Status in Non-Small Cell Lung Cancer. Journal of Respiration. 2025; 5(3):11. https://doi.org/10.3390/jor5030011
Chicago/Turabian StyleBenfares, Anass, Abdelali yahya Mourabiti, Badreddine Alami, Sara Boukansa, Ikram Benomar, Nizar El Bouardi, Moulay Youssef Alaoui Lamrani, Hind El Fatimi, Bouchra Amara, Mounia Serraj, and et al. 2025. "Nomogram Based on the Most Relevant Clinical, CT, and Radiomic Features, and a Machine Learning Model to Predict EGFR Mutation Status in Non-Small Cell Lung Cancer" Journal of Respiration 5, no. 3: 11. https://doi.org/10.3390/jor5030011
APA StyleBenfares, A., Mourabiti, A. y., Alami, B., Boukansa, S., Benomar, I., El Bouardi, N., Alaoui Lamrani, M. Y., El Fatimi, H., Amara, B., Serraj, M., Smahi, M., Cherkaoui, A., Qjidaa, M., Lakhssassi, A., Ouazzani Jamil, M., Maaroufi, M., & Qjidaa, H. (2025). Nomogram Based on the Most Relevant Clinical, CT, and Radiomic Features, and a Machine Learning Model to Predict EGFR Mutation Status in Non-Small Cell Lung Cancer. Journal of Respiration, 5(3), 11. https://doi.org/10.3390/jor5030011






