Differential Effects of Oleuropein and Hydroxytyrosol on Aggregation and Stability of CFTR NBD1-ΔF508 Domain
Abstract
1. Introduction
2. Materials and Methods
2.1. Compounds
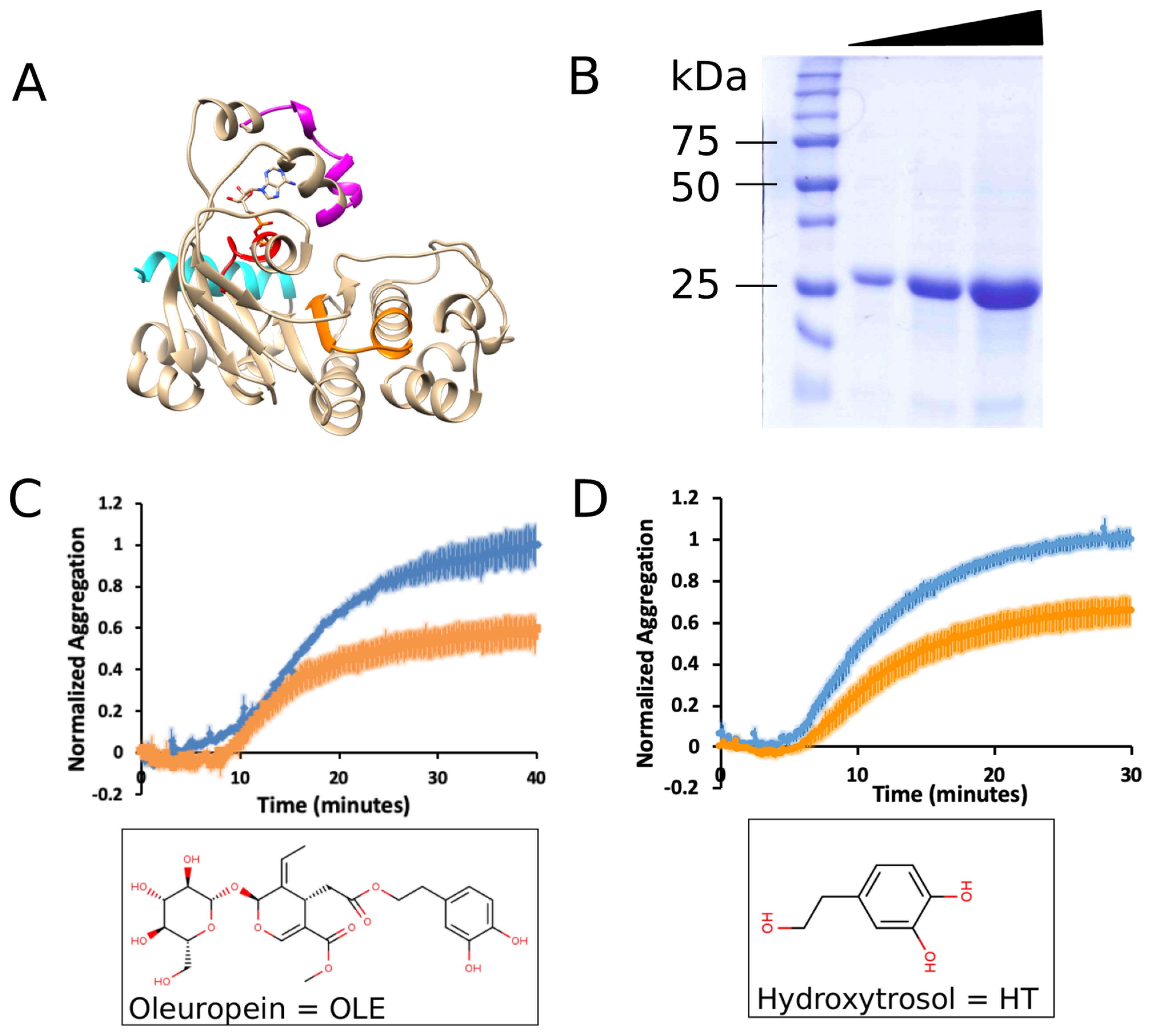
2.2. mNBD1-ΔF508 Expression and Purification
2.3. Aggregation Assay
2.4. Dynamic Light Scattering
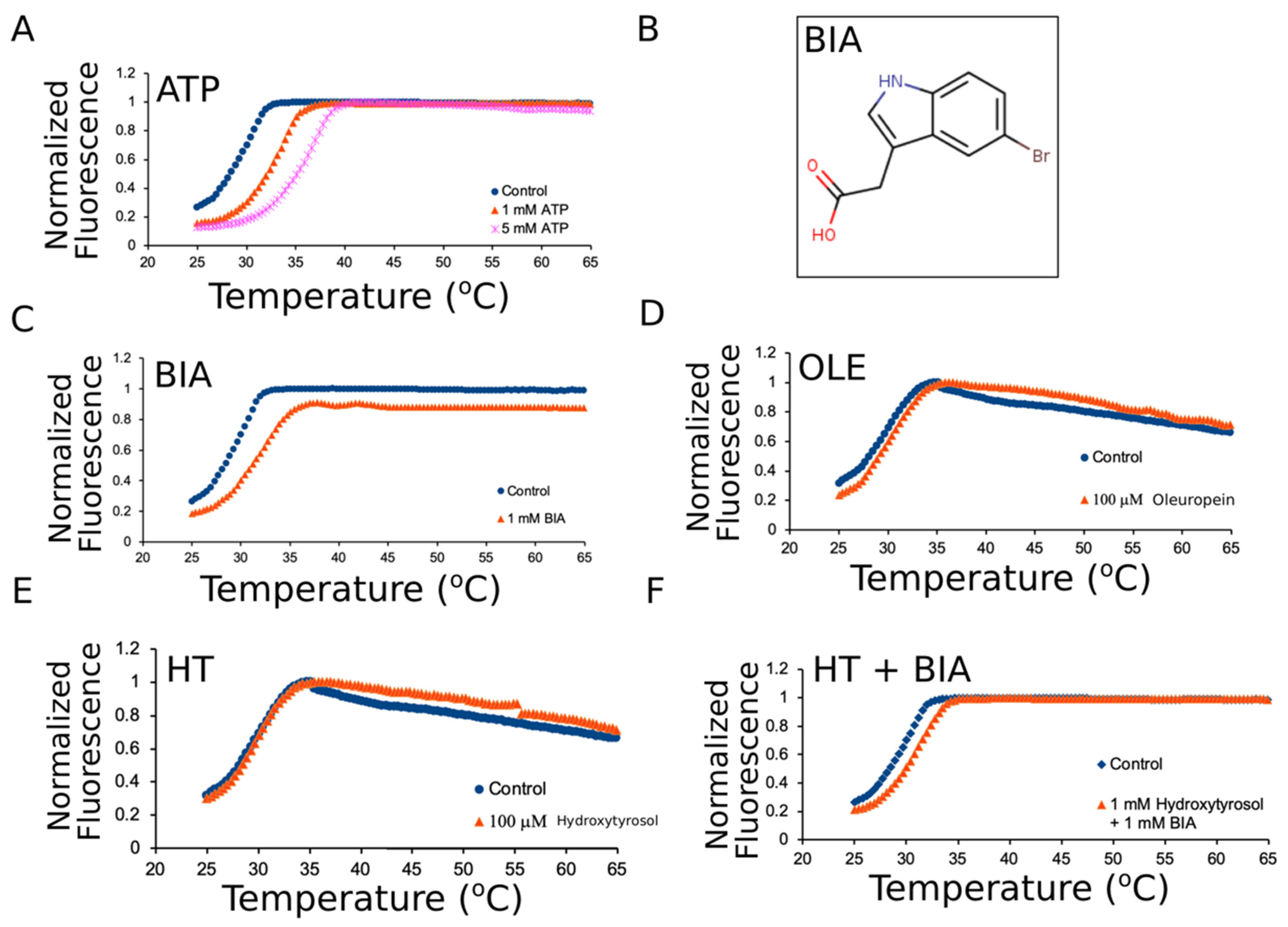
2.5. Differential Scanning Fluorimetry
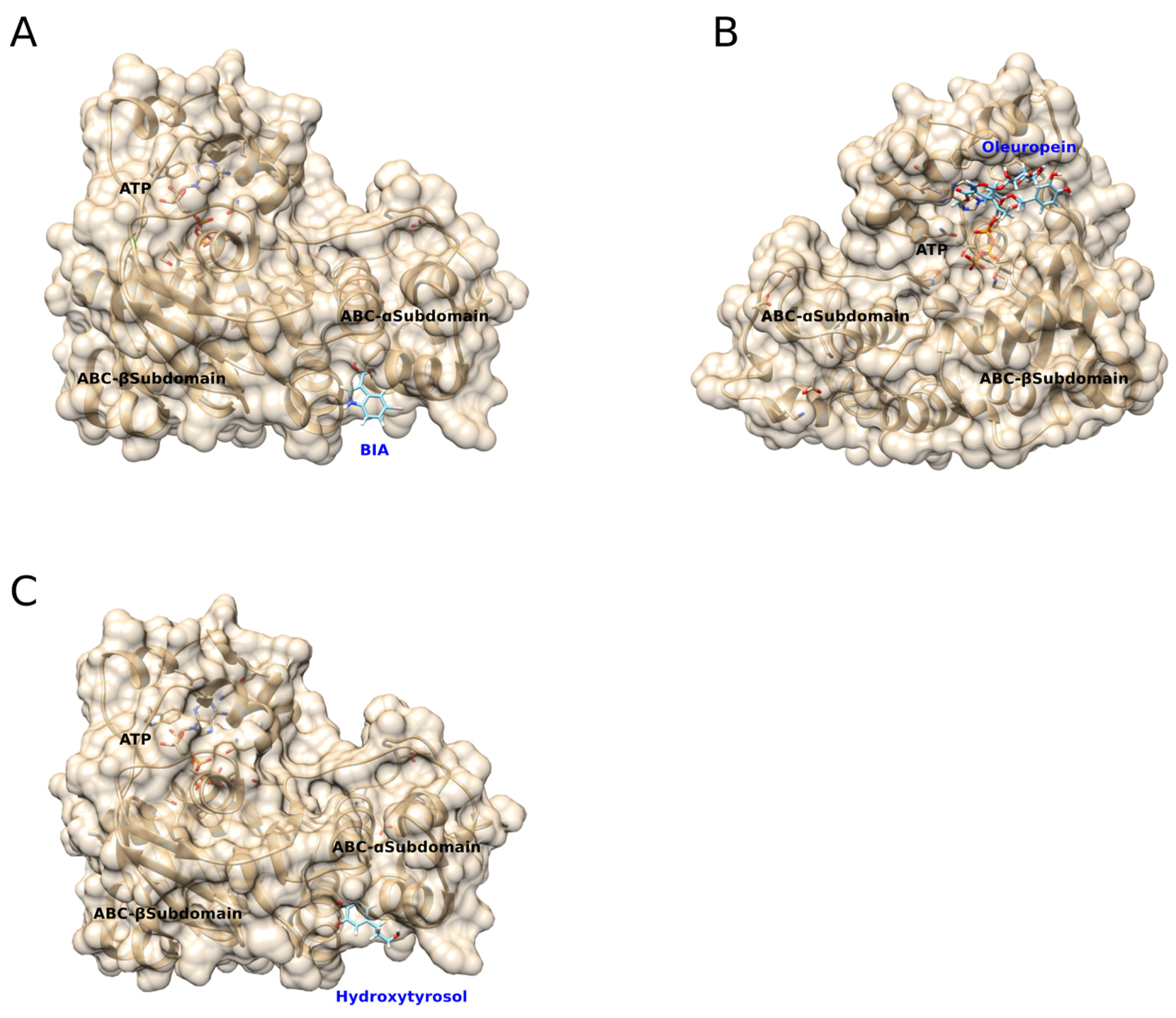
2.6. Molecular Docking Simulations
2.7. Statistical Analysis
3. Results
3.1. Oleuropein and Hydroxytyrosol Suppress the Aggregation of mNBD1-ΔF508
3.2. Oleuropein and Hydroxytyrosol Do Not Thermally Stabilize mNBD1-ΔF508
3.3. Predicted Molecular Docking of BIA, Oleuropein, and Hydroxytryosol to mNBD1-ΔF508
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mense, M.; Vergani, P.; White, D.M.; Altberg, G.; Nairn, A.C.; Gadsby, D.C. In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. EMBO J. 2006, 25, 4728–4739. [Google Scholar] [CrossRef]
- Csanády, L.; Nairn, A.; Gadsby, D.C. Thermodynamics of CFTR Channel Gating: A Spreading Conformational Change Initiates an Irreversible Gating Cycle. J. Gen. Physiol. 2006, 128, 523–533. [Google Scholar] [CrossRef]
- Estabrooks, S.; Brodsky, J.L. Regulation of CFTR Biogenesis by the Proteostatic Network and Pharmacological Modulators. Int. J. Mol. Sci. 2020, 21, 452. [Google Scholar] [CrossRef]
- Riordan, J.R.; Rommens, J.M.; Kerem, B.; Alon, N.; Rozmahel, R.; Grzelczak, Z.; Zielenski, J.; Lok, S.; Plavsic, N.; Chou, J.L.; et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 1989, 245, 1066–1073. [Google Scholar] [CrossRef]
- Lopes-Pacheco, M. CFTR Modulators: Shedding Light on Precision Medicine for Cystic Fibrosis. Front. Pharmacol. 2016, 7, 275. [Google Scholar] [CrossRef] [PubMed]
- The Clinical and Functional TRanslation of CFTR (CFTR2). Available online: http://cftr2.org (accessed on 24 July 2020).
- Castellani, C.; Assael, B.M. Cystic fibrosis: A clinical view. Cell. Mol. Life Sci. 2016, 74, 129–140. [Google Scholar] [CrossRef]
- Denning, G.; Anderson, M.; Amara, J.F.; Marshall, J.; Smith, A.E.; Welsh, M. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 1992, 358, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Teem, J.L.; Berger, H.; Ostedgaard, L.; Rich, D.P.; Tsui, L.-C.; Welsh, M. Identification of revertants for the cystic fibrosis ΔF508 mutation using STE6-CFTR chimeras in yeast. Cell 1993, 73, 335–346. [Google Scholar] [CrossRef]
- Aleksandrov, A.A.; Kota, P.; Aleksandrov, L.A.; He, L.; Jensen, T.; Cui, L.; Gentzsch, M.; Dokholyan, N.V.; Riordan, J.R. Regulatory Insertion Removal Restores Maturation, Stability and Function of ΔF508 CFTR. J. Mol. Biol. 2010, 401, 194–210. [Google Scholar] [CrossRef] [PubMed]
- Farinha, C.; King-Underwood, J.; Sousa, M.; Correia, A.R.; Henriques, B.; Roxo-Rosa, M.; Da Paula, A.C.; Williams, J.; Hirst, S.; Gomes, C.M.; et al. Revertants, Low Temperature, and Correctors Reveal the Mechanism of F508del-CFTR Rescue by VX-809 and Suggest Multiple Agents for Full Correction. Chem. Biol. 2013, 20, 943–955. [Google Scholar] [CrossRef]
- Okiyoneda, T.; Veit, G.; Dekkers, J.F.; Bagdany, M.; Soya, N.; Xu, H.; Roldan, A.; Verkman, A.S.; Kurth, M.J.; Simon, A.; et al. Mechanism-based corrector combination restores ΔF508-CFTR folding and function. Nat. Chem. Biol. 2013, 9, 444–454. [Google Scholar] [CrossRef]
- Sampson, H.M.; Robert, R.; Liao, J.; Matthes, E.; Carlile, G.W.; Hanrahan, J.W.; Thomas, D. Identification of a NBD1-Binding Pharmacological Chaperone that Corrects the Trafficking Defect of F508del-CFTR. Chem. Biol. 2011, 18, 231–242. [Google Scholar] [CrossRef]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Stack, J.H.; Straley, K.S.; Decker, C.J.; Miller, M.; McCartney, J.; Olson, E.R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 2011, 108, 18843–18848. [Google Scholar] [CrossRef]
- Qu, B.-H.; Thomas, P.J. Alteration of the Cystic Fibrosis Transmembrane Conductance Regulator Folding Pathway: Effects of the Δf508 Mutation on the Thermodynamic Stability and Folding Yield of Nbd1. J. Biol. Chem. 1996, 271, 7261–7264. [Google Scholar] [CrossRef]
- Mendoza, J.; Schmidt, A.; Li, Q.; Nuvaga, E.; Barrett, T.; Bridges, R.J.; Feranchak, A.P.; Brautigam, C.; Thomas, P.J. Requirements for Efficient Correction of ΔF508 CFTR Revealed by Analyses of Evolved Sequences. Cell 2012, 148, 164–174. [Google Scholar] [CrossRef]
- Yang, Z.; Hildebrandt, E.; Jiang, F.; Aleksandrov, A.A.; Khazanov, N.; Zhou, Q.; An, J.; Mezzell, A.; Xavier, B.M.; Ding, H.; et al. Structural stability of purified human CFTR is systematically improved by mutations in nucleotide binding domain 1. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Aleksandrov, A.A.; An, J.; Cui, L.; Yang, Z.; Brouillette, C.G.; Riordan, J.R. Restoration of NBD1 Thermal Stability Is Necessary and Sufficient to Correct ∆F508 CFTR Folding and Assembly. J. Mol. Biol. 2015, 427, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Rabeh, W.; Bossard, F.; Xu, H.; Okiyoneda, T.; Bagdany, M.; Mulvihill, C.M.; Du, K.; di Bernardo, S.; Liu, Y.; Konermann, L.; et al. Correction of Both NBD1 Energetics and Domain Interface Is Required to Restore ΔF508 CFTR Folding and Function. Cell 2012, 148, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Micoud, J.; Chauvet, S.; Scheckenbach, K.E.L.; Alfaidy, N.; Chanson, M.; Benharouga, M.; Julien, M. Involvement of the heterodimeric interface region of the nucleotide binding domain-2 (NBD2) in the CFTR quaternary structure and membrane stability. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 2420–2431. [Google Scholar] [CrossRef]
- Carlile, G.W.; Yang, Q.; Matthes, E.; Liao, J.; Radinovic, S.; Miyamoto, C.; Robert, R.; Hanrahan, J.W.; Thomas, D.Y. A novel triple combination of pharmacological chaperones improves F508del-CFTR correction. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Aleksandrov, A.A.; Yang, Z.; Forouhar, F.; Proctor, E.A.; Kota, P.; An, J.; Kaplan, A.; Khazanov, N.; Boël, G.; et al. Ligand binding to a remote site thermodynamically corrects the F508del mutation in the human cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2018, 293, 17685–17704. [Google Scholar] [CrossRef]
- Protasevich, I.; Yang, Z.; Wang, C.; Atwell, S.; Zhao, X.; Emtage, S.; Wetmore, D.; Hunt, J.F.; Brouillette, C.G. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010, 19, 1917–1931. [Google Scholar] [CrossRef]
- Wang, C.; Protasevich, I.; Yang, Z.; Seehausen, D.; Skalak, T.; Zhao, X.; Atwell, S.; Emtage, J.S.; Wetmore, D.R.; Brouillette, C.G.; et al. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci. 2010, 19, 1932–1947. [Google Scholar] [CrossRef]
- Ptitsyn, O.; Pain, R.; Semisotnov, G.; Zerovnik, E.; Razgulyaev, O. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990, 262, 20–24. [Google Scholar] [CrossRef]
- Pissarra, L.S.; Farinha, C.; Xu, Z.; Schmidt, A.; Thibodeau, P.; Cai, Z.; Thomas, P.J.; Sheppard, D.N.; Amaral, M.D. Solubilizing Mutations Used to Crystallize One CFTR Domain Attenuate the Trafficking and Channel Defects Caused by the Major Cystic Fibrosis Mutation. Chem. Biol. 2008, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Marković, A.K.; Torić, J.; Barbarić, M.; Brala, C.J. Hydroxytyrosol, Tyrosol and Derivatives and Their Potential Effects on Human Health. Molecules 2019, 24, 2001. [Google Scholar] [CrossRef]
- Rigacci, S.; Guidotti, V.; Bucciantini, M.; Nichino, D.; Relini, A.; Berti, A.; Stefani, M. Aβ(1-42) Aggregates into Non-Toxic Amyloid Assemblies in the Presence of the Natural Polyphenol Oleuropein Aglycon. Curr. Alzheimer Res. 2011, 8, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Diomede, L.; Rigacci, S.; Romeo, M.; Stefani, M.; Salmona, M. Oleuropein Aglycone Protects Transgenic C. elegans Strains Expressing Aβ42 by Reducing Plaque Load and Motor Deficit. PLoS ONE 2013, 8, e58893. [Google Scholar] [CrossRef]
- Daccache, A.; Lion, C.; Sibille, N.; Gérard, M.; Slomianny, C.; Lippens, G.; Cotelle, P. Oleuropein and derivatives from olives as Tau aggregation inhibitors. Neurochem. Int. 2011, 58, 700–707. [Google Scholar] [CrossRef]
- Youker, R.; Walsh, P.; Beilharz, T.; Lithgow, T.; Brodsky, J.L. Distinct Roles for the Hsp40 and Hsp90 Molecular Chaperones during Cystic Fibrosis Transmembrane Conductance Regulator Degradation in Yeast. Mol. Biol. Cell 2004, 15, 4787–4797. [Google Scholar] [CrossRef]
- Wang, C.K.; Weeratunga, S.; Pacheco, C.M.; Hofmann, A. DMAN: A Java tool for analysis of multi-well differential scanning fluorimetry experiments. Bioinformatics 2011, 28, 439–440. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Strickland, E.; Qu, B.-H.; Millen, L.; Thomas, P.J. The Molecular Chaperone Hsc70 Assists the in VitroFolding of the N-terminal Nucleotide-binding Domain of the Cystic Fibrosis Transmembrane Conductance Regulator. J. Biol. Chem. 1997, 272, 25421–25424. [Google Scholar] [CrossRef] [PubMed]
- Lewis, H.A.; Buchanan, S.G.; Burley, S.K.; Conners, K.; Dickey, M.; Dorwart, M.; Fowler, R.; Gao, X.; Guggino, W.B.; Hendrickson, W.A.; et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2003, 23, 282–293. [Google Scholar] [CrossRef]
- Baker, J.M.R.; Hudson, R.P.; Kanelis, V.; Choy, W.-Y.; Thibodeau, P.H.; Thomas, P.J.; Forman-Kay, J.D. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat. Struct. Mol. Biol. 2007, 14, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Scott-Ward, T.S.; Amaral, M.D. Deletion of Phe508 in the first nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator increases its affinity for the heat shock cognate 70 chaperone: Interaction of Hsc70 with F508del-NBD1. FEBS J. 2009, 276, 7097–7109. [Google Scholar] [CrossRef]
- Hall, J.D.; Wang, H.; Byrnes, L.J.; Shanker, S.; Wang, K.; Efremov, I.V.; Chong, P.A.; Forman-Kay, J.D.; Aulabaugh, A.E. Binding screen for cystic fibrosis transmembrane conductance regulator correctors finds new chemical matter and yields insights into cystic fibrosis therapeutic strategy: CFTR Correctors in CF Therapeutics. Protein Sci. 2016, 25, 360–373. [Google Scholar] [CrossRef][Green Version]
- Atwell, S.; Brouillette, C.G.; Conners, K.; Emtage, S.; Gheyi, T.; Guggino, W.B.; Hendle, J.; Hunt, J.F.; Lewis, H.A.; Lu, F.; et al. Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng. Des. Sel. 2010, 23, 375–384. [Google Scholar] [CrossRef]
- Zapadka, K.L.; Becher, F.J.; Dos Santos, A.L.G.; Jackson, S.E. Factors affecting the physical stability (aggregation) of peptide therapeutics. Interface Focus 2017, 7, 20170030. [Google Scholar] [CrossRef]
- Omar, S.H.; Scott, C.J.; Hamlin, A.S.; Obied, H.K. Olive Biophenols Reduces Alzheimer’s Pathology in SH-SY5Y Cells and APPswe Mice. Int. J. Mol. Sci. 2018, 20, 125. [Google Scholar] [CrossRef]
- Palazzi, L.; Leri, M.; Cesaro, S.; Stefani, M.; Bucciantini, M.; de Laureto, P.P. Insight into the molecular mechanism underlying the inhibition of α-synuclein aggregation by hydroxytyrosol. Biochem. Pharmacol. 2020, 173, 113722. [Google Scholar] [CrossRef] [PubMed]
- Leri, M.; Natalello, A.; Bruzzone, E.; Stefani, M.; Bucciantini, M. Oleuropein aglycone and hydroxytyrosol interfere differently with toxic Aβ1-42 aggregation. Food Chem. Toxicol. 2019, 129, 1–12. [Google Scholar] [CrossRef]
- Sigoillot, M.; Overtus, M.; Grodecka, M.; Scholl, D.; Garcia-Pino, A.; Laeremans, T.; He, L.; Pardon, E.; Hildebrandt, E.; Urbatsch, I.; et al. Domain-interface dynamics of CFTR revealed by stabilizing nanobodies. Nat. Commun. 2019, 10, 2636. [Google Scholar] [CrossRef] [PubMed]
- Kalid, O.; Mense, M.; Fischman, S.; Shitrit, A.; Bihler, H.; Ben-Zeev, E.; Schutz, N.; Pedemonte, N.; Thomas, P.J.; Bridges, R.J.; et al. Small molecule correctors of F508del-CFTR discovered by structure-based virtual screening. J. Comput. Mol. Des. 2010, 24, 971–991. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hudson, R.P.; Dawson, J.E.; Chong, P.A.; Yang, Z.; Millen, L.; Thomas, P.J.; Brouillette, C.G.; Forman-Kay, J.D. Direct Binding of the Corrector VX-809 to Human CFTR NBD1: Evidence of an Allosteric Coupling between the Binding Site and the NBD1:CL4 Interface. Mol. Pharmacol. 2017, 92, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Bitam, S.; Elbahnsi, A.; Creste, G.; Pranke, I.; Chevalier, B.; Berhal, F.; Hoffmann, B.; Servel, N.; Tondelier, D.; Hatton, A.; et al. New insights into structure and function of bis-phosphinic acid derivatives and implications for CFTR modulation. Sci. Rep. 2021, 11, 6842. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, L.J.; Xu, Y.; Qiu, X.; Hall, J.D.; West, G.M. Sites associated with Kalydeco binding on human Cystic Fibrosis Transmembrane Conductance Regulator revealed by Hydrogen/Deuterium Exchange. Sci. Rep. 2018, 8, 4664. [Google Scholar] [CrossRef]
| Groups | Average °C | Variance |
|---|---|---|
| mNBD1-ΔF508 | 29.97 | 0.32 |
| mNBD1-ΔF508 + 1 mM ATP | 33.70 | 0.64 |
| mNBD1-ΔF508 + 5 mM ATP | 36.13 | 0.05 |
| mNBD1-ΔF508 + 1 mM BIA | 32.10 | 0.16 |
| mNBD1-ΔF508 + 1.5 mM ATP + 1 mM BIA | 35.87 | 0.21 |
| mNBD1-ΔF508 + 100 μM Oleuropein | 30.23 | 0.40 |
| mNBD1-ΔF508 + 100 μM Hydroxy-Tyrosol | 29.83 | 0.40 |
| mNBD1-ΔF508 + 500 μM Hydroxy-Tyrosol | 29.73 | 0.16 |
| mNBD1-ΔF508 + 1 mM Hydroxy-Tyrosol | 29.47 | 0.56 |
| mNBD1-ΔF508 + 1 mM Hydroxy-Tyrosol + 1 mM BIA | 31.53 | 0.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, C.S.; Wyderko, J.A.; Vang, Y.; Martin, G.; Youker, R.T. Differential Effects of Oleuropein and Hydroxytyrosol on Aggregation and Stability of CFTR NBD1-ΔF508 Domain. J. Respir. 2021, 1, 204-215. https://doi.org/10.3390/jor1030019
Robinson CS, Wyderko JA, Vang Y, Martin G, Youker RT. Differential Effects of Oleuropein and Hydroxytyrosol on Aggregation and Stability of CFTR NBD1-ΔF508 Domain. Journal of Respiration. 2021; 1(3):204-215. https://doi.org/10.3390/jor1030019
Chicago/Turabian StyleRobinson, Christopher S., Jennifer A. Wyderko, Yeng Vang, Galen Martin, and Robert T. Youker. 2021. "Differential Effects of Oleuropein and Hydroxytyrosol on Aggregation and Stability of CFTR NBD1-ΔF508 Domain" Journal of Respiration 1, no. 3: 204-215. https://doi.org/10.3390/jor1030019
APA StyleRobinson, C. S., Wyderko, J. A., Vang, Y., Martin, G., & Youker, R. T. (2021). Differential Effects of Oleuropein and Hydroxytyrosol on Aggregation and Stability of CFTR NBD1-ΔF508 Domain. Journal of Respiration, 1(3), 204-215. https://doi.org/10.3390/jor1030019





