Delving into the Role of Receptor-like Tyrosine Kinase (RYK) in Cancer: In Silico Insights into Its Diagnostic and Prognostic Utility
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of RYK Expression in Cancer-Derived RNA Data Sets
2.2. RYK Expression in Cancer Subtypes RNA Data Sets
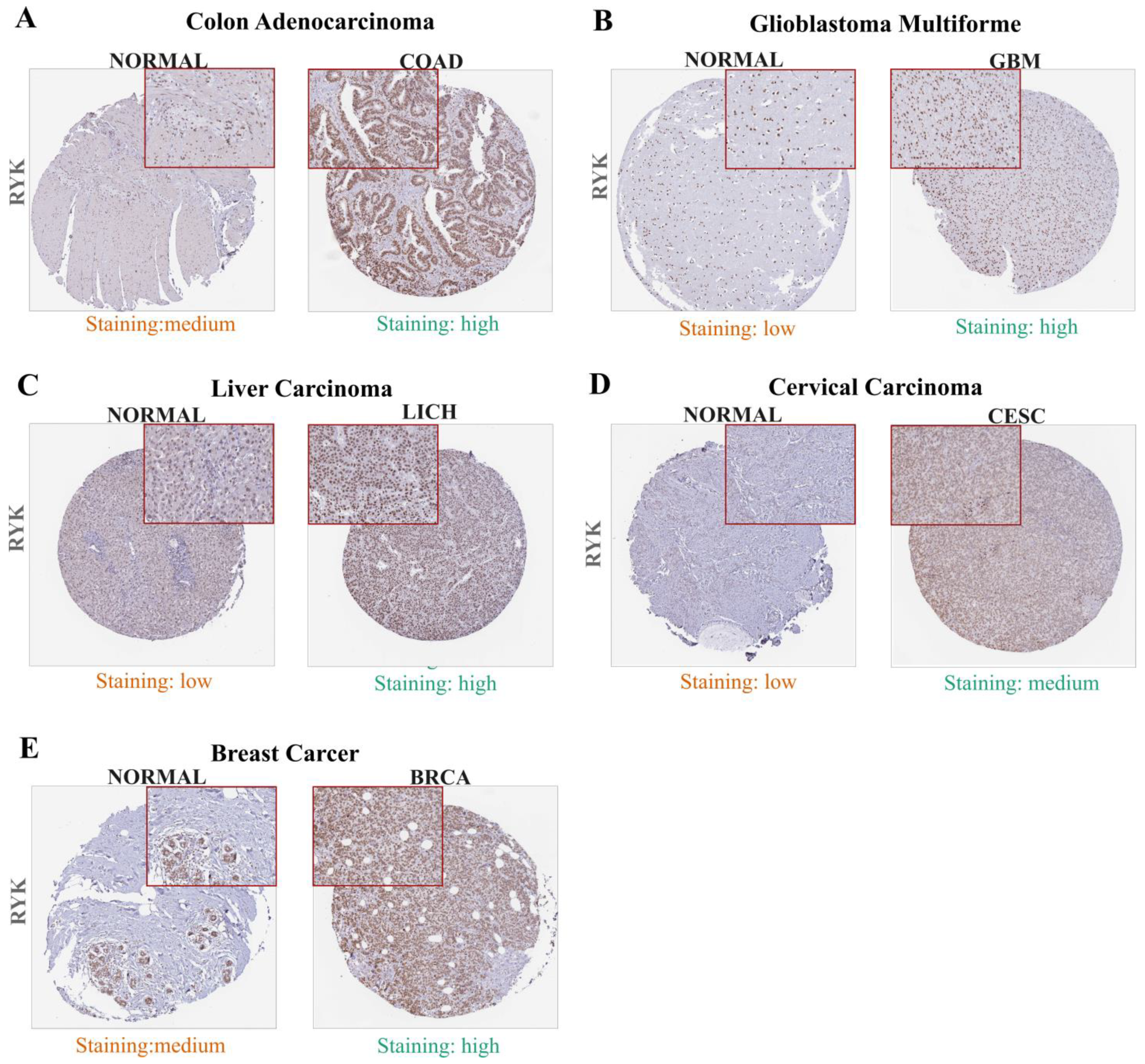
2.3. Protein Expression Analysis
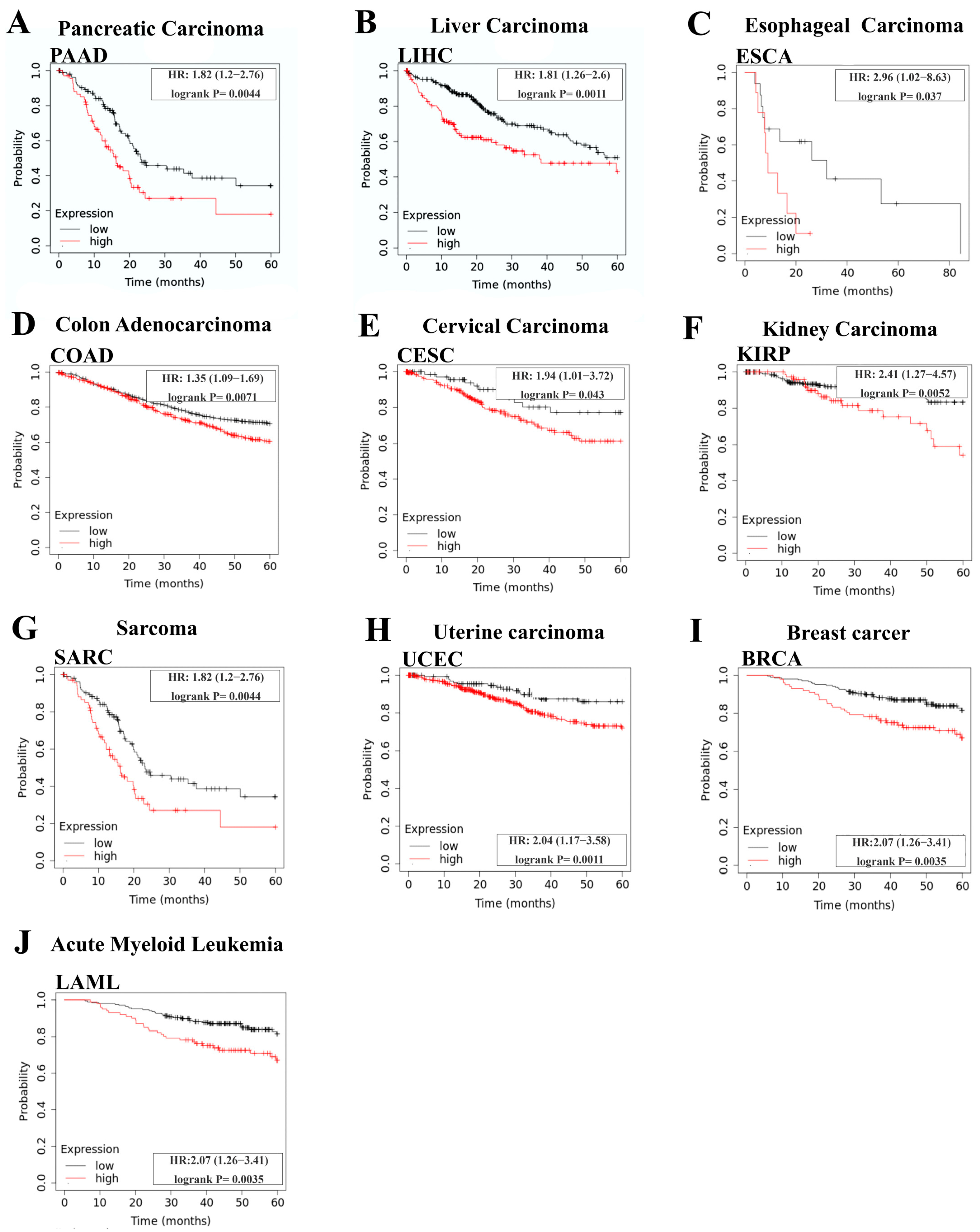
2.4. Survival Analysis
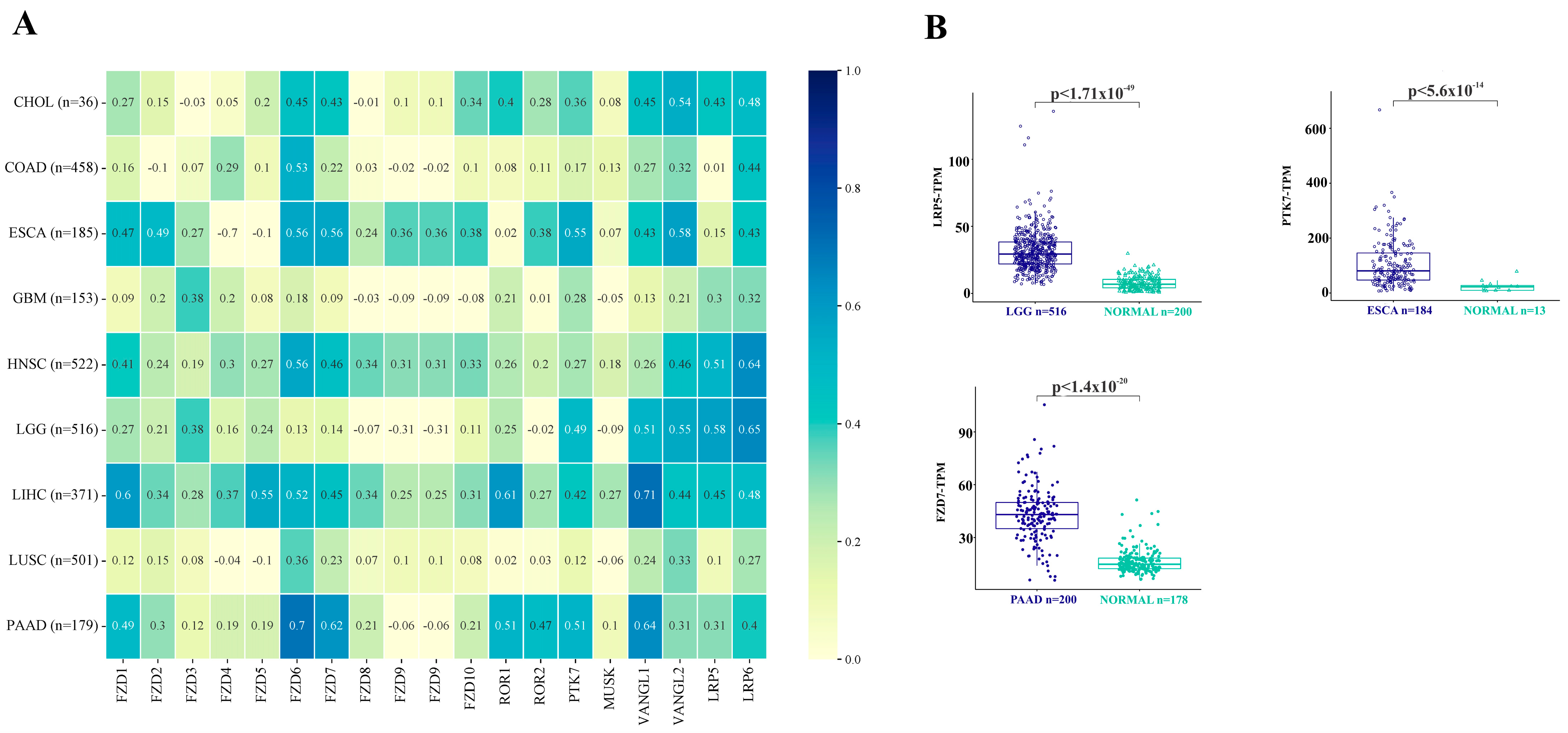
2.5. Correlation and Expression of WNT Pathway Receptors and Ligands
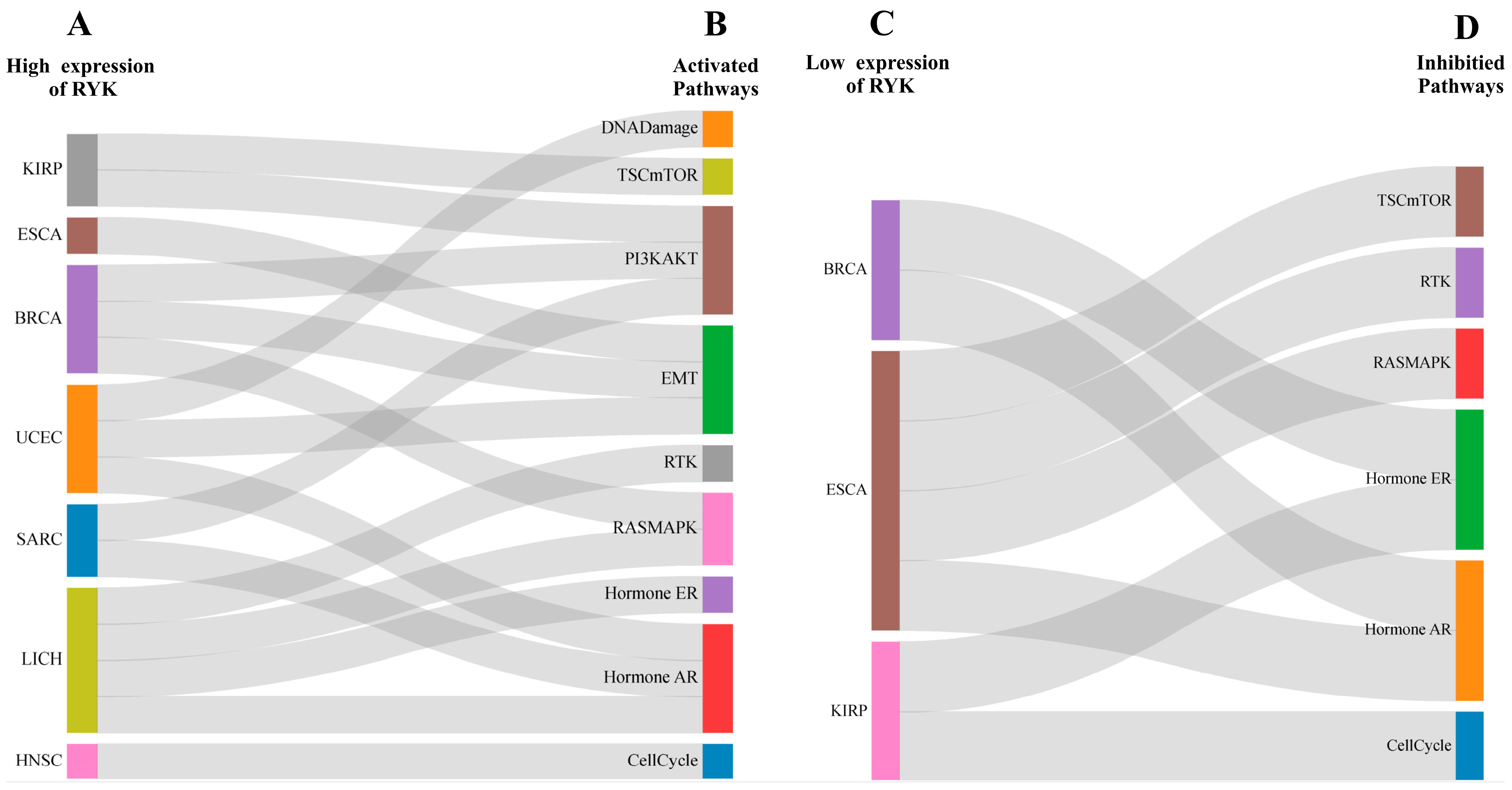
2.6. Expression and Relation of RYK with Cancer-Related Pathways
3. Results
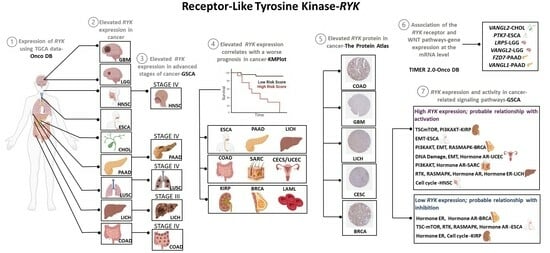
3.1. RYK Overexpression in Cancer
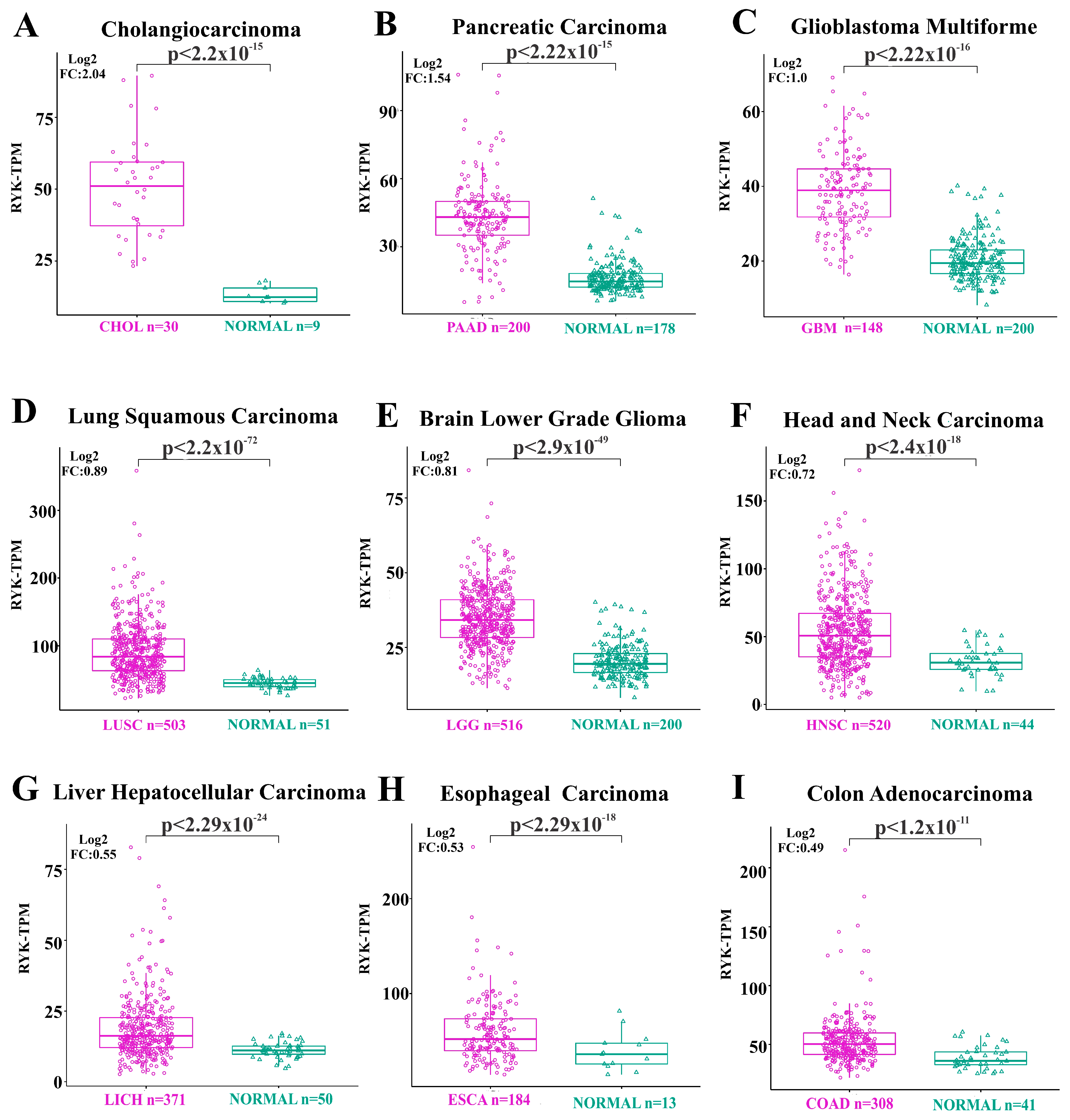
3.2. Overexpression of Receptor-like Tyrosine Kinase in Cancer- and Non-Cancer-Derived Tissues
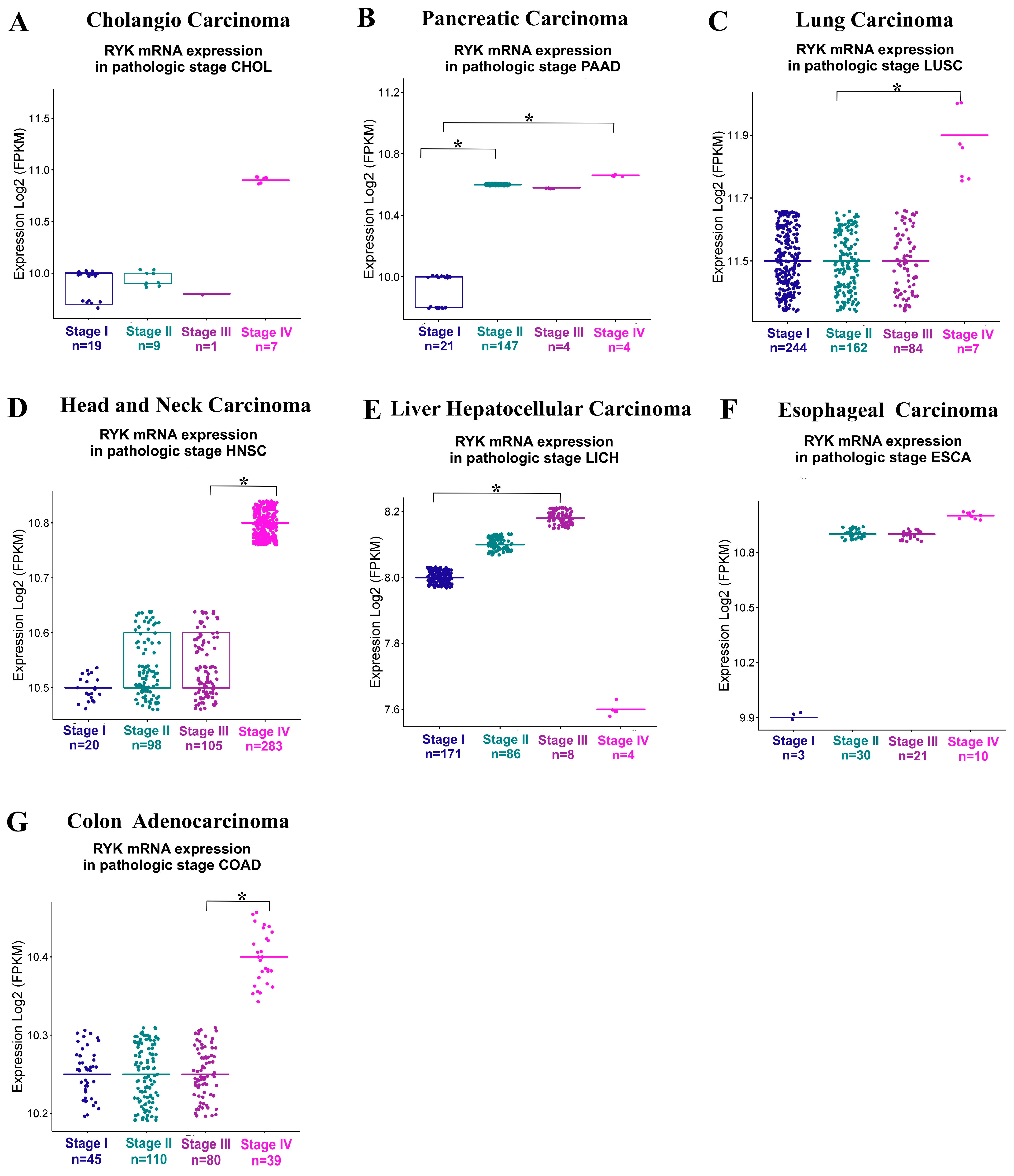
3.3. Prognostic Value of RYK in Cancer
3.4. Correlation of the WNT Signaling Pathway Receptors and Ligands with RYK Expression in Cancer- and Non-Cancer-Derived Samples
3.5. Association of RYK with Ten Cancer-Related Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halford, M.M.; Macheda, M.L.; Stacker, S.A. The RYK Receptor Family. In Receptor Tyrosine Kinases: Family and Subfamilies; Springer: Cham, Switzerland, 2015; pp. 685–741. Available online: https://link.springer.com/chapter/10.1007/978-3-319-11888-8_15 (accessed on 11 November 2023).
- Hovens, C.M.; Stacker, S.A.; Andres, A.C.; Harpur, A.G.; Ziemiecki, A.; Wilks, A.F. RYK, a receptor tyrosine kinase-related molecule with unusual kinase domain motifs. Proc. Natl. Acad. Sci. USA 1992, 89, 11818–11822. [Google Scholar] [CrossRef]
- Green, J.; Nusse, R.; van Amerongen, R. The role of Ryk and Ror receptor tyrosine kinases in Wnt signal transduction. Cold Spring Harb. Perspect. Biol. 2014, 6, a009175. [Google Scholar] [CrossRef]
- Roy, J.P.; Halford, M.M.; Stacker, S.A. The biochemistry, signalling and disease relevance of RYK and other WNT-binding receptor tyrosine kinases. Growth Factors 2018, 36, 15–40. [Google Scholar] [CrossRef]
- Bovolenta, P.; Rodriguez, J.; Esteve, P. Frizzled/RYK mediated signalling in axon guidance. Development 2006, 133, 4399–4408. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, S.; Xu, Y.; Zhao, C. Regulation of ABCG2 expression by Wnt5a through FZD7 in human pancreatic cancer cells. Mol. Med. Rep. 2021, 23, 52. [Google Scholar] [CrossRef] [PubMed]
- Bie, J.; Hu, X.; Yang, M.; Shi, X.; Zhang, X.; Wang, Z. PTK7 promotes the malignant properties of cancer stem-like cells in esophageal squamous cell lines. Hum. Cell 2020, 33, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Song, G.; Zhang, X.; Li, Q.; Zhao, Y.; Zhou, Y.; Xiong, R.; Hu, X.; Tang, Z.; Feng, G. PTK7 is a novel oncogenic target for esophageal squamous cell carcinoma. World J. Surg. Oncol. 2017, 15, 105. [Google Scholar] [CrossRef][Green Version]
- Macheda, M.L.; Sun, W.W.; Kugathasan, K.; Hogan, B.M.; Bower, N.I.; Halford, M.M.; Zhang, Y.F.; Jacques, B.E.; Lieschke, G.J.; Dabdoub, A.; et al. The Wnt receptor Ryk plays a role in mammalian planar cell polarity signaling. J. Biol. Chem. 2012, 287, 29312–29323. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Zavala, M.; Riveros-Magana, A.R.; Garcia-Castro, B.; Barrera-Chairez, E.; Rubio-Jurado, B.; Garces-Ruiz, O.M.; Ramos-Solano, M.; Aguilar-Lemarroy, A.; Jave-Suarez, L.F. WNT receptors profile expression in mature blood cells and immature leukemic cells: RYK emerges as a hallmark receptor of acute leukemia. Eur. J. Haematol. 2016, 97, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Halford, M.M.; Macheda, M.L.; Parish, C.L.; Takano, E.A.; Fox, S.; Layton, D.; Nice, E.; Stacker, S.A. A fully human inhibitory monoclonal antibody to the Wnt receptor RYK. PLoS ONE 2013, 8, e75447. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Lu, C.C.; Kerman, R.; Steward, O.; Xu, X.M.; Zou, Y. Repulsive Wnt signaling inhibits axon regeneration after CNS injury. J. Neurosci. 2008, 28, 8376–8382. [Google Scholar] [CrossRef]
- Tury, A.; Tolentino, K.; Zou, Y. Altered expression of atypical PKC and Ryk in the spinal cord of a mouse model of amyotrophic lateral sclerosis. Dev. Neurobiol. 2014, 74, 839–850. [Google Scholar] [CrossRef]
- Ventura, E.; Belfiore, A.; Iozzo, R.V.; Giordano, A.; Morrione, A. Progranulin and EGFR modulate receptor-like tyrosine kinase sorting and stability in mesothelioma cells. Am. J. Physiol. Cell Physiol. 2023, 325, C391–C405. [Google Scholar] [CrossRef]
- Hamilton, G.; Rath, B.; Klameth, L.; Hochmair, M. Receptor tyrosine kinase expression of circulating tumor cells in small cell lung cancer. Oncoscience 2015, 2, 629–634. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, Y.; Huang, J.; Cai, Z.; Wang, Y. RYK, a receptor of noncanonical Wnt ligand Wnt5a, is positively correlated with gastric cancer tumorigenesis and potential of liver metastasis. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G352–G360. [Google Scholar] [CrossRef]
- Habu, M.; Koyama, H.; Kishida, M.; Kamino, M.; Iijima, M.; Fuchigami, T.; Tokimura, H.; Ueda, M.; Tokudome, M.; Koriyama, C.; et al. Ryk is essential for Wnt-5a-dependent invasiveness in human glioma. J. Biochem. 2014, 156, 29–38. [Google Scholar] [CrossRef]
- Adamo, A.; Fiore, D.; De Martino, F.; Roscigno, G.; Affinito, A.; Donnarumma, E.; Puoti, I.; Ricci Vitiani, L.; Pallini, R.; Quintavalle, C.; et al. RYK promotes the stemness of glioblastoma cells via the WNT/ beta-catenin pathway. Oncotarget 2017, 8, 13476–13487. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Liu, L.; Gao, R.; Che, C.; Yang, G. Downregulation of exosomal miR-7-5p promotes breast cancer migration and invasion by targeting RYK and participating in the atypical WNT signalling pathway. Cell. Mol. Biol. Lett. 2022, 27, 88. [Google Scholar] [CrossRef] [PubMed]
- Ohara, S.; Suda, K.; Fujino, T.; Hamada, A.; Koga, T.; Nishino, M.; Chiba, M.; Shimoji, M.; Takemoto, T.; Soh, J.; et al. Dose-dependence in acquisition of drug tolerant phenotype and high RYK expression as a mechanism of osimertinib tolerance in lung cancer. Lung Cancer 2021, 154, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Okura, R.; Fujihara, S.; Iwama, H.; Morishita, A.; Chiyo, T.; Watanabe, M.; Hirose, K.; Kobayashi, K.; Fujimori, T.; Kato, K.; et al. MicroRNA profiles during galectin-9-induced apoptosis of pancreatic cancer cells. Oncol. Lett. 2018, 15, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Reddy, R.B.; Khora, S.S.; Suresh, A. Molecular prognosticators in clinically and pathologically distinct cohorts of head and neck squamous cell carcinoma-A meta-analysis approach. PLoS ONE 2019, 14, e0218989. [Google Scholar] [CrossRef]
- Tang, G.; Cho, M.; Wang, X. OncoDB: An interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022, 50, D1334–D1339. [Google Scholar] [CrossRef]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Kassambara, A. Package ‘ggpubr’. 2013. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 1 November 2023).
- Ross Ihaka, R.G. R Programming Language. 1993. Available online: https://cran.r-project.org/bin/windows/base/old/ (accessed on 25 October 2023).
- Jung, K.; Panko, P.; Lee, J.; Hwang, H. A Comparative Study on the Performance of GSCA and CSA in Parameter Recovery for Structural Equation Models with Ordinal Observed Variables. Front. Psychol. 2018, 9, 2461. [Google Scholar] [CrossRef]
- Liu, C.J.; Hu, F.F.; Xie, G.Y.; Miao, Y.R.; Li, X.W.; Zeng, Y.; Guo, A.Y. GSCA: An integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief. Bioinform. 2023, 24, bbac558. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheat Maps Package. 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 10 October 2023).
- Akbani, R.; Ng, P.K.; Werner, H.M.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.Y.; Yoshihara, K.; Li, J.; et al. Corrigendum: A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2015, 6, 4852. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; Posit, P.B.C. ggplot2. 2023. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 25 October 2023).
- Bojanowski, M. Alluvial Diagrams. 2019. Available online: https://cran.r-project.org/web/packages/alluvial/vignettes/alluvial.html (accessed on 25 October 2023).
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J. Receptor tyrosine kinases: Legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 2014, 6, a008912. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018, 62, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Lojk, J.; Marc, J. Roles of Non-Canonical Wnt Signalling Pathways in Bone Biology. Int. J. Mol. Sci. 2021, 22, 10840. [Google Scholar] [CrossRef] [PubMed]
- Duchartre, Y.; Kim, Y.M.; Kahn, M. The Wnt signaling pathway in cancer. Crit. Rev. Oncol. Hematol. 2016, 99, 141–149. [Google Scholar] [CrossRef]
- Parsons, M.J.; Tammela, T.; Dow, L.E. WNT as a Driver and Dependency in Cancer. Cancer Discov. 2021, 11, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Micci, F.; Panagopoulos, I.; Haugom, L.; Andersen, H.K.; Tjonnfjord, G.E.; Beiske, K.; Heim, S. t(3;21)(q22;q22) leading to truncation of the RYK gene in atypical chronic myeloid leukemia. Cancer Lett. 2009, 277, 205–211. [Google Scholar] [CrossRef]
- He, W.; Yuan, K.; He, J.; Wang, C.; Peng, L.; Han, Y.; Chen, N. Network and pathway-based analysis of genes associated with esophageal squamous cell carcinoma. Ann. Transl. Med. 2023, 11, 102. [Google Scholar] [CrossRef]
- Zhou, G.; Zhang, S.; Jin, M.; Hu, S. Comprehensive analysis reveals COPB2 and RYK associated with tumor stages of larynx squamous cell carcinoma. BMC Cancer 2022, 22, 667. [Google Scholar] [CrossRef]
- Lyu, J.; Wesselschmidt, R.L.; Lu, W. Cdc37 regulates Ryk signaling by stabilizing the cleaved Ryk intracellular domain. J. Biol. Chem. 2009, 284, 12940–12948. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Greenbaum, D.; Colangelo, C.; Williams, K.; Gerstein, M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Skoda, J.; Neradil, J.; Staniczkova Zambo, I.; Nunukova, A.; Macsek, P.; Borankova, K.; Dobrotkova, V.; Nemec, P.; Sterba, J.; Veselska, R. Serial Xenotransplantation in NSG Mice Promotes a Hybrid Epithelial/Mesenchymal Gene Expression Signature and Stemness in Rhabdomyosarcoma Cells. Cancers 2020, 12, 196. [Google Scholar] [CrossRef] [PubMed]
- Ueno, K.; Hirata, H.; Hinoda, Y.; Dahiya, R. Frizzled homolog proteins, microRNAs and Wnt signaling in cancer. Int. J. Cancer 2013, 132, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Halford, M.M.; Armes, J.; Buchert, M.; Meskenaite, V.; Grail, D.; Hibbs, M.L.; Wilks, A.F.; Farlie, P.G.; Newgreen, D.F.; Hovens, C.M.; et al. Ryk-deficient mice exhibit craniofacial defects associated with perturbed Eph receptor crosstalk. Nat. Genet. 2000, 25, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Karimi Roshan, M.; Soltani, A.; Soleimani, A.; Rezaie Kahkhaie, K.; Afshari, A.R.; Soukhtanloo, M. Role of AKT and mTOR signaling pathways in the induction of epithelial-mesenchymal transition (EMT) process. Biochimie 2019, 165, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Kang, M.; Chang, C.; Wei, H.; Zhang, C.; Chen, Y. Multiple forms of cell death: A focus on the PI3K/AKT pathway. J. Cell. Physiol. 2023, 238, 2026–2038. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, X. Crosstalk between Wnt/beta-catenin signaling pathway and DNA damage response in cancer: A new direction for overcoming therapy resistance. Front. Pharmacol. 2023, 14, 1230822. [Google Scholar] [CrossRef] [PubMed]
- Persad, A.; Venkateswaran, G.; Hao, L.; Garcia, M.E.; Yoon, J.; Sidhu, J.; Persad, S. Active beta-catenin is regulated by the PTEN/PI3 kinase pathway: A role for protein phosphatase PP2A. Genes. Cancer 2016, 7, 368–382. [Google Scholar] [CrossRef] [PubMed]
- Trivier, E.; Ganesan, T.S. RYK, a catalytically inactive receptor tyrosine kinase, associates with EphB2 and EphB3 but does not interact with AF-6. J. Biol. Chem. 2002, 277, 23037–23043. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapata-García, J.A.; Jave-Suárez, L.F.; Aguilar-Lemarroy, A. Delving into the Role of Receptor-like Tyrosine Kinase (RYK) in Cancer: In Silico Insights into Its Diagnostic and Prognostic Utility. J. Mol. Pathol. 2024, 5, 66-80. https://doi.org/10.3390/jmp5010005
Zapata-García JA, Jave-Suárez LF, Aguilar-Lemarroy A. Delving into the Role of Receptor-like Tyrosine Kinase (RYK) in Cancer: In Silico Insights into Its Diagnostic and Prognostic Utility. Journal of Molecular Pathology. 2024; 5(1):66-80. https://doi.org/10.3390/jmp5010005
Chicago/Turabian StyleZapata-García, Jessica Alejandra, Luis Felipe Jave-Suárez, and Adriana Aguilar-Lemarroy. 2024. "Delving into the Role of Receptor-like Tyrosine Kinase (RYK) in Cancer: In Silico Insights into Its Diagnostic and Prognostic Utility" Journal of Molecular Pathology 5, no. 1: 66-80. https://doi.org/10.3390/jmp5010005
APA StyleZapata-García, J. A., Jave-Suárez, L. F., & Aguilar-Lemarroy, A. (2024). Delving into the Role of Receptor-like Tyrosine Kinase (RYK) in Cancer: In Silico Insights into Its Diagnostic and Prognostic Utility. Journal of Molecular Pathology, 5(1), 66-80. https://doi.org/10.3390/jmp5010005