Use of the Biocartis Idylla™ Platform for the Detection of Epidermal Growth Factor Receptor, BRAF and KRAS Proto-Oncogene Mutations in Liquid-Based Cytology Specimens from Patients with Non-Small Cell Lung Carcinoma and Pancreatic Adenocarcinoma
Abstract
:1. Introduction
Objectives
2. Methods
Preparation of Samples
3. Results
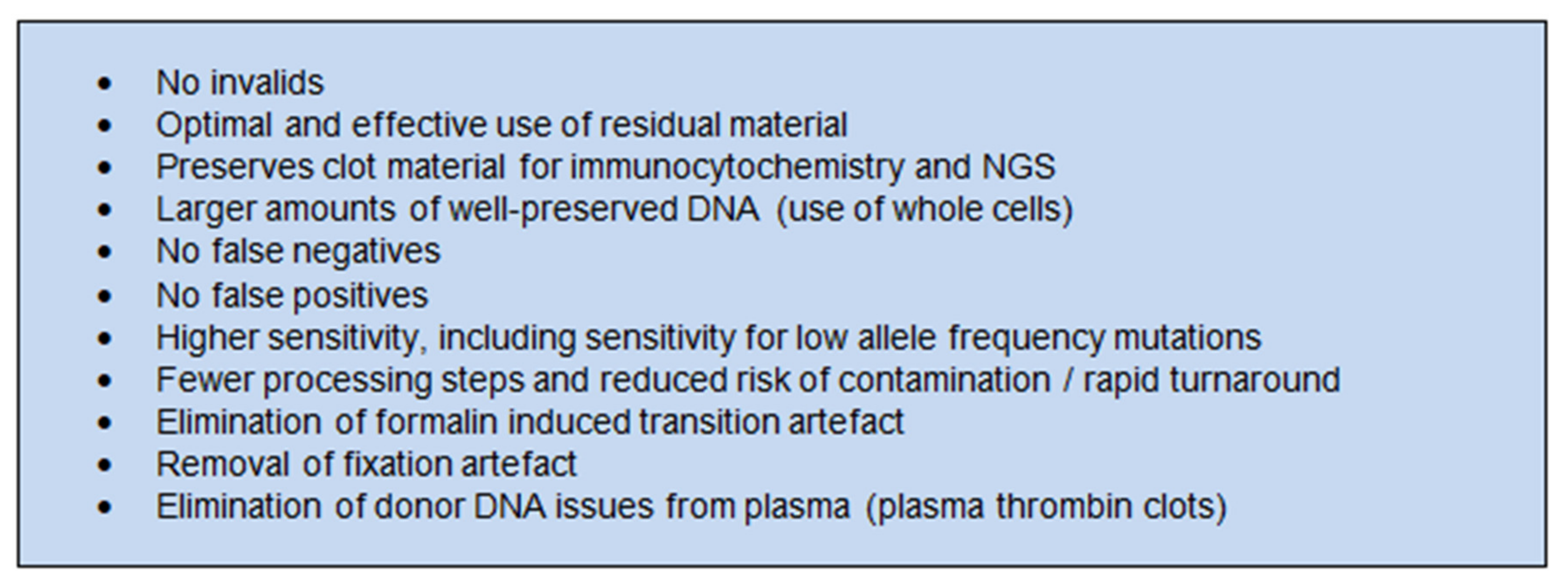
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Research UK. 2021 Lung Cancer Statistics Based on: Cancer Incidence from Cancer Intelligence Statistical Information Team at Cancer Research UK (201520142017 UK Average). Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics-for-the-uk (accessed on 17 December 2021).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Office for National Statistics. Cancer Survival by Stage at Diagnosis for England. Available online: https://www.gov.uk/government/statistics/cancer-survival-in-england-for-patients-diagnosed-between-2014-and-2018-and-followed-up-until-2019/cancer-survival-in-england-for-patients-diagnosed-between-2014-and-2018-and-followed-up-to-2019 (accessed on 12 August 2021).
- National Comprehensive Cancer Network. NCCN Guidelines: Non-Small Cell Lung Cancer. Version 1.2020. 2019. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (accessed on 8 January 2020).
- Liam, C.K.; Mallawathantri, S.; Fong, K.M. Is tissue still the issue in detecting molecular alterations in lung cancer? Respirology 2020, 25, 933–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NICE guideline [NG122] Lung Cancer: Diagnosis and Management. Available online: https://www.nice.org.uk/guidance/ng122 (accessed on 9 November 2021).
- Wood, K.; Hensing, T.; Malik, R.; Salgia, R. Prognostic and Predictive Value inKRASin Non–Small-Cell Lung Cancer. JAMA Oncol. 2016, 2, 805–812. [Google Scholar] [CrossRef]
- Arbour, K.C.; Jordan, E.; Kim, H.R.; Dienstag, J.; Yu, H.A.; Sanchez-Vega, F.; Lito, P.; Berger, M.; Solit, D.B.; Hellmann, M.; et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non–Small Cell Lung Cancer. Clin. Cancer Res. 2017, 24, 334–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheffler, M.; Ihle, M.A.; Hein, R.; Merkelbach-Bruse, S.; Scheel, A.H.; Siemanowski, J.; Brägelmann, J.; Kron, A.; Abedpour, N.; Ueckeroth, F.; et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019, 14, 606–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.T.; Solomon, B. Targeting Anaplastic Lymphoma Kinase in Lung Cancer. Clin. Cancer Res. 2011, 17, 2081–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, L.H.; Souza, B.M.; Leite, L.R.; Parma, S.A.F.; Lopes, N.P.; Malta, F.S.V.; Freire, M.C.M. Molecular profile of KRAS G12C-mutant colorectal and non-small-cell lung cancer. BMC Cancer 2021, 21, 193. [Google Scholar] [CrossRef]
- Amgen. Amgen’s Investigational KRAS G12C Inhibitor Sotorasib Demonstrated Rapid, Deep and Durable Responses in Previously Treated Patients with Advanced Non-Small Cell Lung Cancer. Available online: https://prn.to/3px7knl (accessed on 28 January 2021).
- Sotorasib–Medicines–SPS–Specialist Pharmacy Service–The First Stop for Professional Medicines Advice. Available online: https://www.sps.nhs.uk/medicines/sotorasib/ (accessed on 28 January 2021).
- Lee, M.S.; Pant, S. Personalizing Medicine with Germline and Somatic Sequencing in Advanced Pancreatic Cancer: Current Treatments and Novel Opportunities. Am. Soc. Clin. Oncol. Educ. Book 2021, 41, e153–e165. [Google Scholar] [CrossRef]
- Kenney, C.; Kunst, T.; Webb, S.; Christina, D.; Arrowood, C.; Steinberg, S.M.; Mettu, N.B.; Kim, E.J.; Rudloff, U. Phase II study of selumetinib, an orally active inhibitor of MEK1 and MEK2 kinases, in KRASG12R-mutant pancreatic ductal adenocarcinoma. Investig. New Drugs 2021, 39, 821–828. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Bengtsson, A.; Andersson, R.; Ansari, D. The actual 5-year survivors of pancreatic ductal adenocarcinoma based on real-world data. Sci. Rep. 2020, 10, 16425. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 153–168. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.-C.; Mansour, J.; Mollaee, M.; Wagner, K.-U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef]
- Du Rand, I.A.; Barber, P.V.; Goldring, J.; Lewis, R.A.; Mandal, S.; Munavvar, M.; Rintoul, R.; Shah, P.; Singh, S.; Slade, M.G.; et al. Summary of the British Thoracic Society Guidelines for advanced diagnostic and therapeutic flexible bronchoscopy in adults. Thorax 2011, 66, 1014–1015. [Google Scholar] [CrossRef] [Green Version]
- Shetty, D.; Bhatnagar, G.; Sidhu, H.S.; Fox, B.M.; Dodds, N.I. The increasing role of endoscopic ultrasound (EUS) in the man-agement of pancreatic and biliary disease. Clin. Radiol. 2013, 68, 323–335. [Google Scholar] [CrossRef]
- Tian, S.K.; Killian, J.K.; Rekhtman, N.; Benayed, R.; Middha, S.; Ladanyi, M.; Lin, O.; Arcila, M.E. Optimizing Workflows and Processing of Cytologic Samples for Comprehensive Analysis by Next-Generation Sequencing: Memorial Sloan Kettering Cancer Center Experience. Arch. Pathol. Lab. Med. 2016, 140, 1200–1205. [Google Scholar] [CrossRef] [Green Version]
- Arcila, M.E.; Yang, S.-R.; Momeni, A.; Mata, D.A.; Salazar, P.; Chan, R.; Elezovic, D.; Benayed, R.; Zehir, A.; Buonocore, D.J.; et al. Ultrarapid EGFR Mutation Screening Followed by Comprehensive Next-Generation Sequencing: A Feasible, Informative Approach for Lung Carcinoma Cytology Specimens with a High Success Rate. JTO Clin. Res. Rep. 2020, 1, 100077. [Google Scholar] [CrossRef]
- Petiteau, C.; Robinet-Zimmermann, G.; Riot, A.; Dorbeau, M.; Richard, N.; Blanc-Fournier, C.; Bibeau, F.; Deshayes, S.; Bergot, E.; Gervais, R.; et al. Contribution of the IdyllaTM System to Improving the Therapeutic Care of Patients with NSCLC through Early Screening of EGFR Mutations. Curr. Oncol. 2021, 28, 4432–4445. [Google Scholar] [CrossRef]
- Evrard, S.M.; Taranchon-Clermont, E.; Rouquette, I.; Murray, S.; Dintner, S.; Nam-Apostolopoulos, Y.-C.; Bellosillo, B.; Varela, M.; Nadal, E.; Wiedorn, K.H.; et al. Multicenter Evaluation of the Fully Automated PCR-Based Idylla EGFR Mutation Assay on Formalin-Fixed, Paraffin-Embedded Tissue of Human Lung Cancer. J. Mol. Diagn. 2019, 21, 1010–1024. [Google Scholar] [CrossRef]
- Glinski, L.; Shetty, D.; Iles, S.; Diggins, B.; Garvican, J. Single slide assessment: A highly effective cytological rapid on-site evaluation technique for endobronchial and endoscopic ultrasound-guided fine needle aspiration. Cytopathology 2018, 30, 164–172. [Google Scholar] [CrossRef]
- Pisapia, P.; Pepe, F.; Sgariglia, R.; Nacchio, M.; Russo, G.; Conticelli, F.; Girolami, I.; Eccher, A.; Bellevicine, C.; Vigliar, E.; et al. Next generation sequencing in cytology. Cytopathology 2021, 32, 588–595. [Google Scholar] [CrossRef]
- Finall, A.; Davies, G.; Jones, T.; Emlyn, G.; Huey, P.; Mullard, A. Integration of rapid PCR testing as an adjunct to NGS in diagnostic pathology services within the UK: Evidence from a case series of non-squamous, non-small cell lung cancer (NSCLC) patients with follow-up. J. Clin. Pathol. 2022, 18, 2021–207987. [Google Scholar] [CrossRef]
- Gregg, J.P.; Li, T.; Yoneda, K.Y. Molecular testing strategies in non-small cell lung cancer: Optimizing the diagnostic journey. Transl. Lung Cancer Res. 2019, 8, 286–301. [Google Scholar] [CrossRef] [Green Version]

| KRAS | EGFR | BRAF | TOTAL | |
|---|---|---|---|---|
| PANCREATIC ADENOCARCINOMA | 8 | - | - | 8 |
| LUNG ADENOCARCINOMA | 8 | 29 | 8 | 45 |
| LUNG SQUAMOUS CELL CARCINOMA | 6 | - | - | 6 |
| TOTAL | 22 | 29 | 8 * | 59 |
| Study Number | LBC Slide Tumour % | LBC Result | LBC WT Cq Value | LBC Mutation Cq Value | LBC Delta Cq Value | Clot Slide Tumour % | Clot Result | Clot WT Cq Value | Clot Mutation Cq Value | Clot Delta Cq Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Panc 1 | 5% >50 cells | KRAS Q61H mutation detected | 21.74 | 31.68 | 9.73 | <100 cells <5% tumour | KRAS Q61H mutation detected | 25.39 | 29.93 | 4.35 |
| Panc 2 | 5% >100 cells | KRAS Q61H mutation detected | 22.35 | 30.12 | 7.65 | 100 cells 10% tumour | KRAS Q61H mutation detected | 25.53 | 29.23 | 3.8 |
| Panc 3 | 50% >100 cells | KRAS G12D mutation detected | 25.15 | 28.03 | 3.04 | <100 cells 5% tumour | KRAS G12D mutation detected | 25.45 | 30.85 | 5.67 |
| Panc 5 A | Slide not found | KRAS G12R mutation detected | 19.8 | 23.41 | 3.59 | >100 cells 60% tumour | KRAS G12R mutation detected | 24.30 | 31.12 | 6.86 |
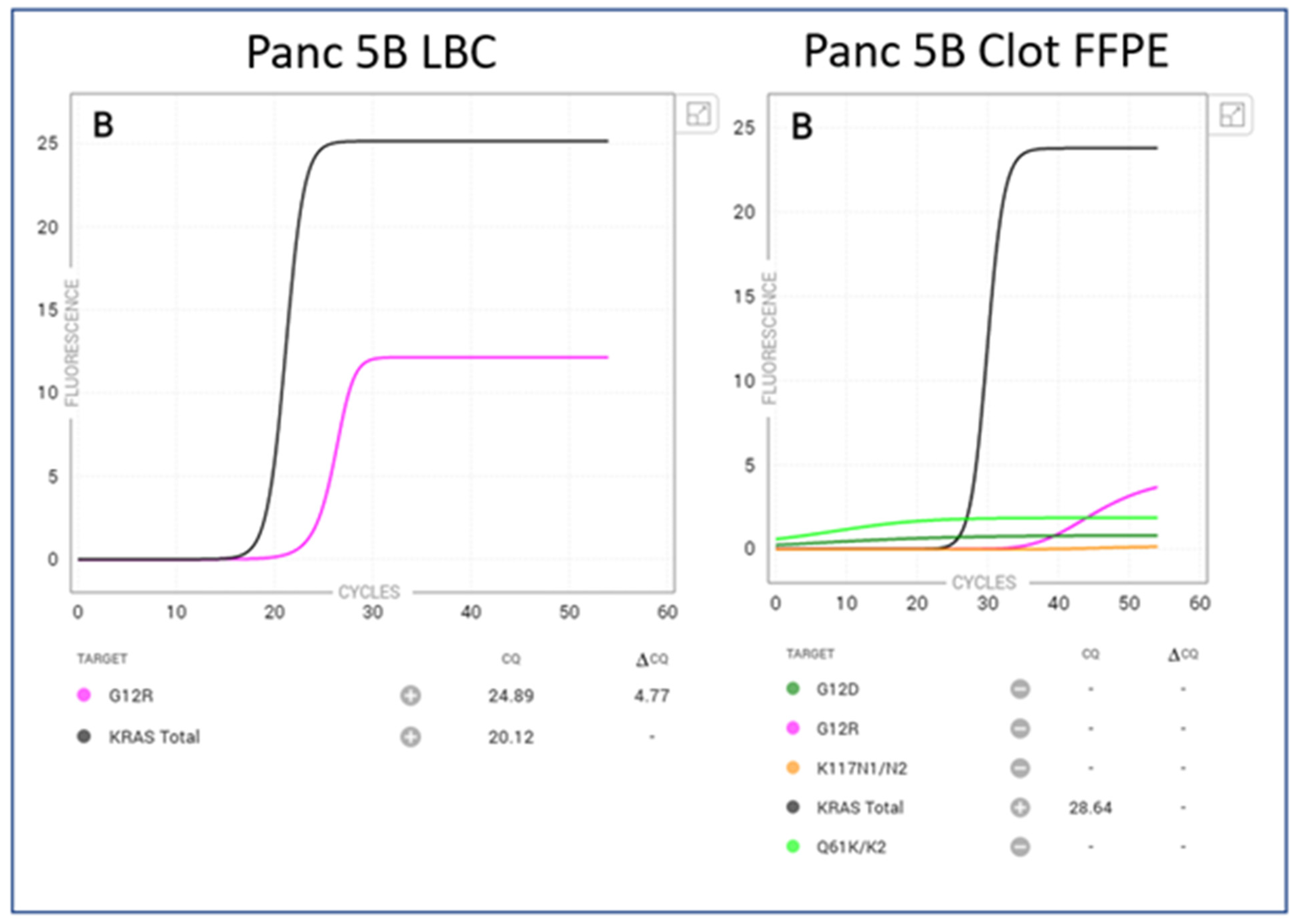
| Panc 5 B | 40% >100 cells | KRAS G12R mutation detected | 20.24 | 24.89 | 4.77 | <50 cells 5% tumour | No mutation detected | 28.36 | n/a | n/a |
| Panc 6 A | 10% >100 cells | KRAS G12D mutation detected | 18.9 | 22.9 | 4.05 | >100cells 80% tumour | KRAS G12D mutation detected | 21.64 | 25.39 | 3.75 |
| Panc 6 B | 10% >100 cells | KRAS G12D mutation detected | 18.37 | 21.34 | 3.03 | <100 cells <5% tumour | KRAS G12D mutation detected | 25.39 | 29.93 | 4.35 |
| Lung adeno L021 | 40%>20 cells | KRAS G12Cmutation detected | 22.41 | 27.61 | 6.24 | <200 cells >50% tumour | KRAS G12Cmutation detected | 23.8 | 28.08 | 5.00 |
| Lung SCC L038 | 90% >100 cells | KRAS G12V mutation detected | 18.21 | 26.72 | 8.44 | >200 cells >80% | No mutation detected | 24.55 | n/a | n/a |
| Lung adeno L002 | 50% >200 cells | EGFR Mutation detected Exon 19 | 15.4 | 20 | 4.87 | 80% 500 cells | EGFR Mutation detected Exon 19 | 21 | 26 | 5.66 |
| Lung adeno L012 | 30% >200 cells | EGFR Mutation detected Exon 19 deletion | 20.7 | 25.2 | 4.18 | 80% 100 cells. | EGFR Mutation detected Exon 19 | 21.5 | 26.32 | 4.62 |
| Lung adeno L015 | 90% >200 cells | No mutation detected | 18.8 | n/a | n/a | 90%. >200 | EGFR Mutation detected S768I * false positive | 26.1 | 28.24 | 2.94 |
| Lung adeno L044 | 5%>100 cells | EGFR Mutation detected Exon 19 | 19.15 | 27.38 | 7.85 | 75% >200 cells | EGFR Mutation detected Exon 19 | 25.2 | 29.01 | 3.80 |
| Total Tests | Number of Mutations Detected | Invalids LBC | Invalids Clot FFPE | |
|---|---|---|---|---|
| KRAS LBC | 22 | 9 | 0/22 (0%) | 0/22 (0%) |
| KRAS Clot | 22 | 7 | ||
| EFGR LBC | 29 | 3 | 0/29 (0%) | 0/29 (0%) |
| EGFR Clot | 29 | 3 | ||
| BRAF LBC | 8 | 0 | 0/8 (0%) | 4/8 (50%) |
| BRAF Clot | 8 | 0 |
| Test | Neoplastic Cell Content (LBC PAP) | Pellet Volume | WT Cq Value | Delta Cq | Valid Amplification Curve |
|---|---|---|---|---|---|
| KRAS | 5%>50 cells | 20 μL | <33 | Low |  |
| EGFR | 5%>100 cells | 20 μL | <26 | Low |  |
| BRAF | >50% | 20 μL | <33 *IFU | Low |  |
| LBC Average Cq Value | Clot FFPE Average Cq Value | LBC Average Mutation Cq Value | Clot FFPE Average Mutation Cq Value | |
|---|---|---|---|---|
| EGFR | 17.6 | 22.4 | 24.1 | 27.3 |
| KRAS | 21.4 | 24.5 | 25.8 | 28.7 |
| BRAF | 30.9 | 34.29 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wheeldon, L.; Jones, M.; Probyn, B.; Shetty, D.; Garvican, J. Use of the Biocartis Idylla™ Platform for the Detection of Epidermal Growth Factor Receptor, BRAF and KRAS Proto-Oncogene Mutations in Liquid-Based Cytology Specimens from Patients with Non-Small Cell Lung Carcinoma and Pancreatic Adenocarcinoma. J. Mol. Pathol. 2022, 3, 104-114. https://doi.org/10.3390/jmp3020010
Wheeldon L, Jones M, Probyn B, Shetty D, Garvican J. Use of the Biocartis Idylla™ Platform for the Detection of Epidermal Growth Factor Receptor, BRAF and KRAS Proto-Oncogene Mutations in Liquid-Based Cytology Specimens from Patients with Non-Small Cell Lung Carcinoma and Pancreatic Adenocarcinoma. Journal of Molecular Pathology. 2022; 3(2):104-114. https://doi.org/10.3390/jmp3020010
Chicago/Turabian StyleWheeldon, Leonie, Mary Jones, Ben Probyn, Dushyant Shetty, and James Garvican. 2022. "Use of the Biocartis Idylla™ Platform for the Detection of Epidermal Growth Factor Receptor, BRAF and KRAS Proto-Oncogene Mutations in Liquid-Based Cytology Specimens from Patients with Non-Small Cell Lung Carcinoma and Pancreatic Adenocarcinoma" Journal of Molecular Pathology 3, no. 2: 104-114. https://doi.org/10.3390/jmp3020010
APA StyleWheeldon, L., Jones, M., Probyn, B., Shetty, D., & Garvican, J. (2022). Use of the Biocartis Idylla™ Platform for the Detection of Epidermal Growth Factor Receptor, BRAF and KRAS Proto-Oncogene Mutations in Liquid-Based Cytology Specimens from Patients with Non-Small Cell Lung Carcinoma and Pancreatic Adenocarcinoma. Journal of Molecular Pathology, 3(2), 104-114. https://doi.org/10.3390/jmp3020010






