Comparison of Capability of SAR and Optical Data in Mapping Forest above Ground Biomass Based on Machine Learning †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Set
2.2.1. Field Data
2.2.2. Remote Sensing Data
- Optical Data
- SAR Data
2.3. Methodology
- (1)
- processing of remotely sensed data which includes preprocessing, feature extraction (calculation of vegetation indices, tasseled cap, texture parameters and principal component analysis (PCA) of Optical data, and extraction of Polarimetric decompositions, texture characteristics and backscatter coefficients of SAR data);
- (2)
- selection of the optimal features, generating a biomass estimation model, and biomass map generation.
2.3.1. Preprocessing of Remote Sensing Data
- Optical Data
- SAR Data
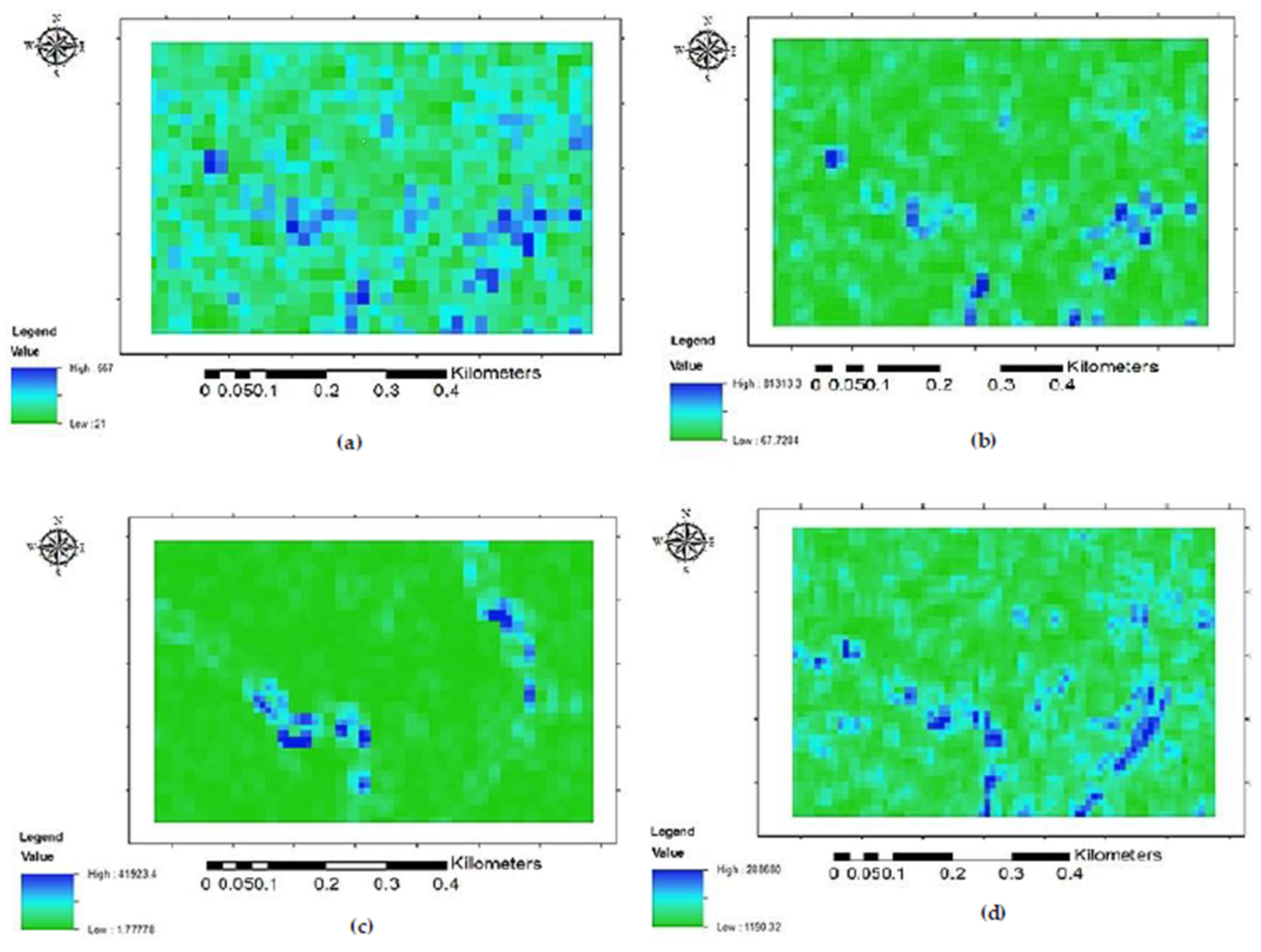
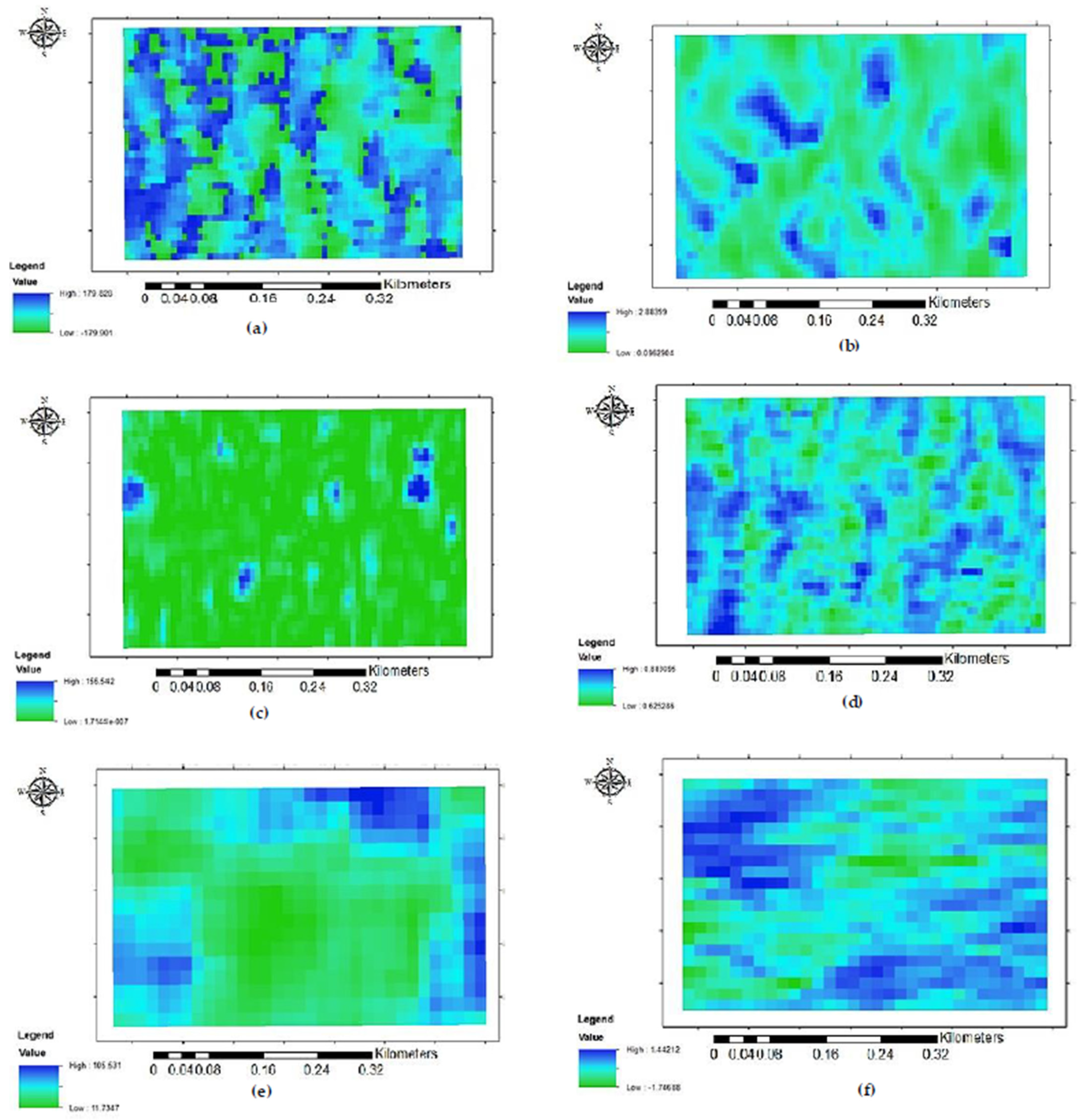
2.3.2. Feature Extraction
- Optical Feature Extraction
- (1)
- Vegetation Index (VI): VI is some mathematical constituent or spectral bands transform that shows the spectral properties of plants which appear distinct from other image features. Vegetation information from remote sensing data is mainly interpreted by the differences and variations of green leaves of plants and spectral features of the canopy [36]. Vegetation index enhances plant signal while reducing solar irradiance and soil background effects [37].The vegetation indices used are described in Table 1 [38,39,40,41].
- (2)
- Texture: The texture is related to the spatial distribution of the intensity values in the image and the grayscale characteristics and expresses the spatial distribution of the pixel values in the image. Therefore, the texture can be described as the spatial distribution of grayscale in a neighborhood. Texture plays an important role in image analysis and pattern recognition. A number of texture features are extracted from the GLCM. The GLCM method is a way for extracting texture properties based on second-order statistics [42].
- (3)
- Tasseled Cap: the tasseled cap is a transformation for converting the original image bands into a new set of bands with defined interpretations useful for vegetation mapping. The transformation is performed to evaluate the change in green biomass based on three components which are brightness, greenness, and wetness, indicating the correlation of the visible and infrared bands. Brightness and greenness shows changes in soil reflectance and variations in the power of green plants, respectively and wetness indicates surface moisture [43,44].
- (4)
- Principle Component Analysis (PCA): PCA can identify the main components and help us to analyze a set of features that have more information instead of all features. PCA is widely used to remove waste data in satellite data. Principal component analysis is divided into three steps; The first step is to obtain the variance-covariance matrix, the second step is to calculate the eigenvectors, and the third step is to linearly transform the data set [45].
- Radar Feature Extraction
- (1)
- Back Scatter Coefficient: The normalized measure of returned radar signal from a distributed target is called the back scatter coefficient and is defined as the unit of the surface. The back scatter depends on the dielectric content properties of the surface. Analysis and evaluation of backscatter coefficients can provide valuable information on surface moisture content, surface roughness and dielectric content, and vegetation cover [46]. The back scatter of a canopy depends on the geometry of the leaves, branches, trunk, and moisture inside the canopy. Back scatter of the forest may include a number of scattering components from different parts of the forest, such as volume scattering of canopy branches, back scatter of the ground, and back scatter of branches and leaves [47,48].
- (2)
- Polarimetric Decomposition: Polarimetric decomposition is the decomposition of the coherent matrix or covariance matrix [47] into a set of independent matrices that exhibit independent scattering related to various physical scattering mechanisms such as surface, double bounce and volume scattering. The Polarimetric decomposition of SAR data is an analysis to determine different types of backscatter. There are different types of decomposition methods. The Polarimetric decomposition methods used are m-sigma decomposition, m-chi (m_) decomposition, m-alpha (m_α) decomposition [49,50,51,52], compact decomposition [53], Eigen vector decomposition [47] and H/A/Alpha decomposition [54].
- (3)
- Texture: The texture feature of radar data is the same as the texture feature of optical data.
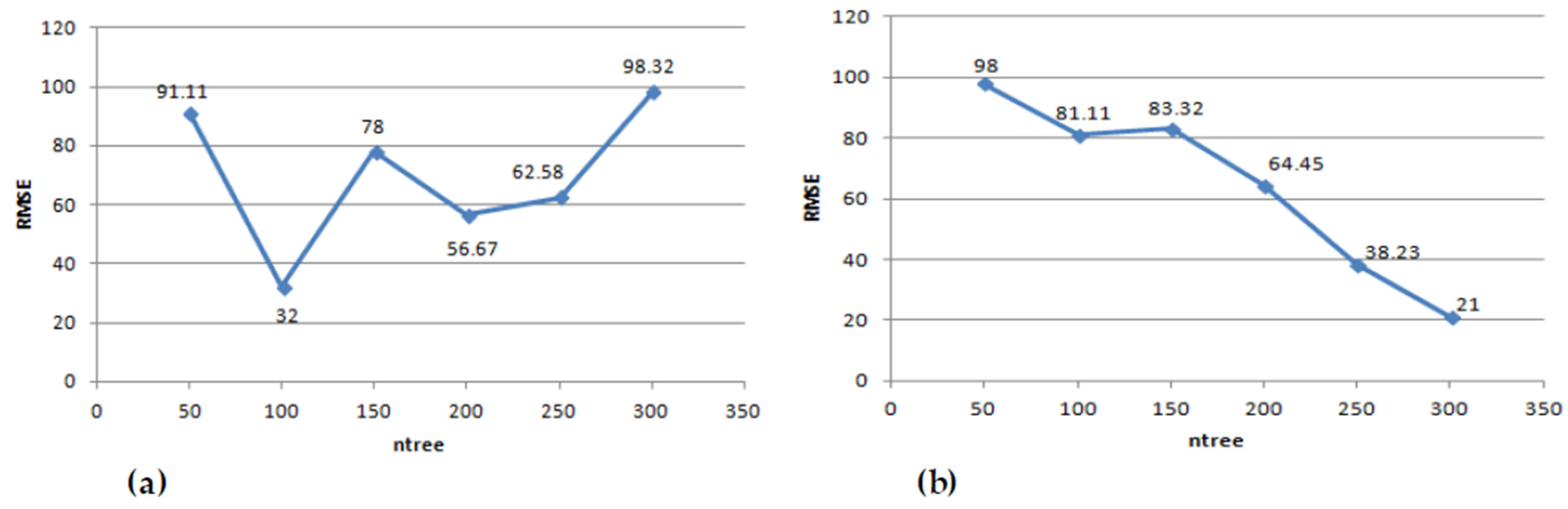
2.3.3. Above Ground Biomass Modelling Based on GA-RF
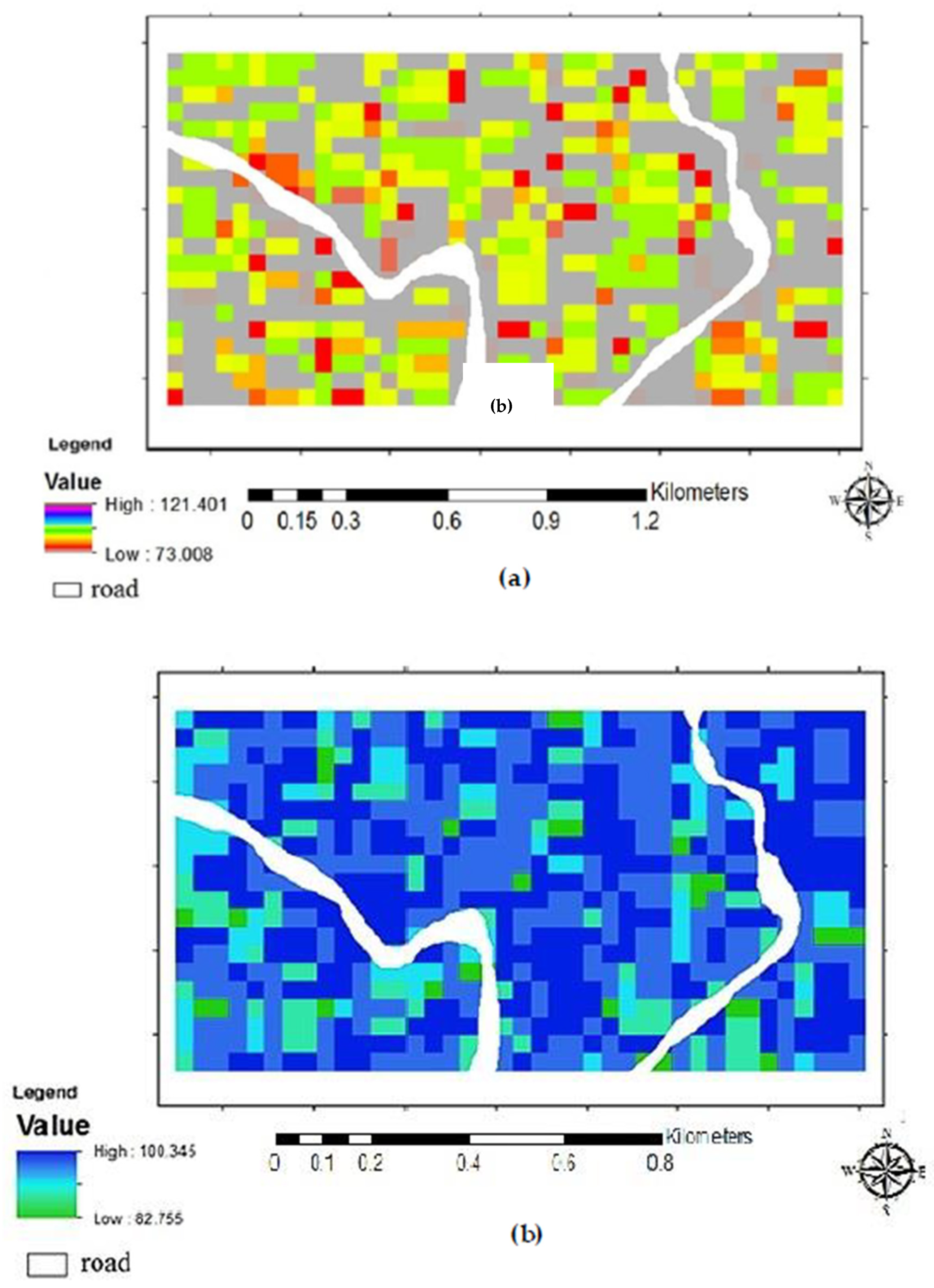
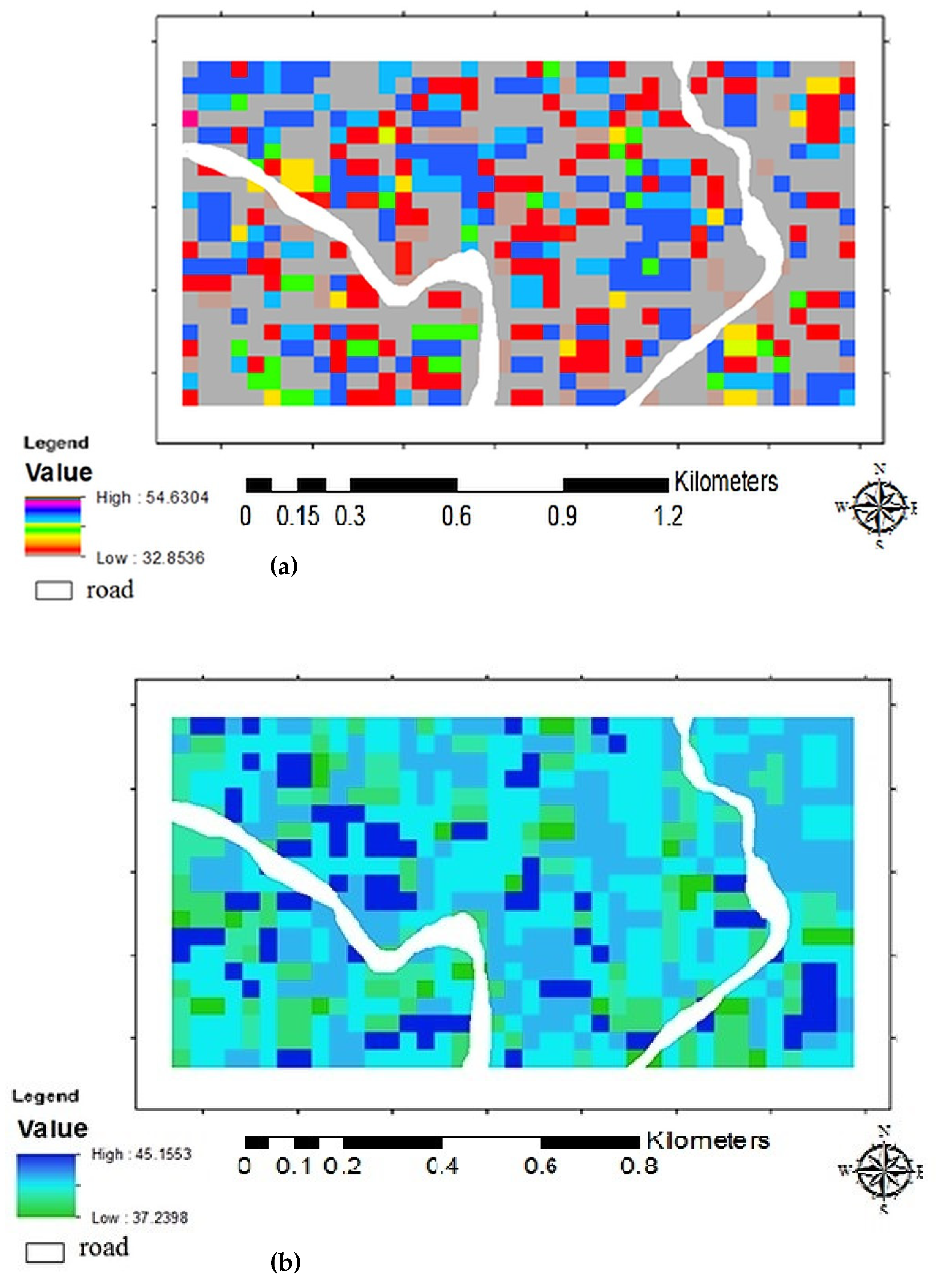
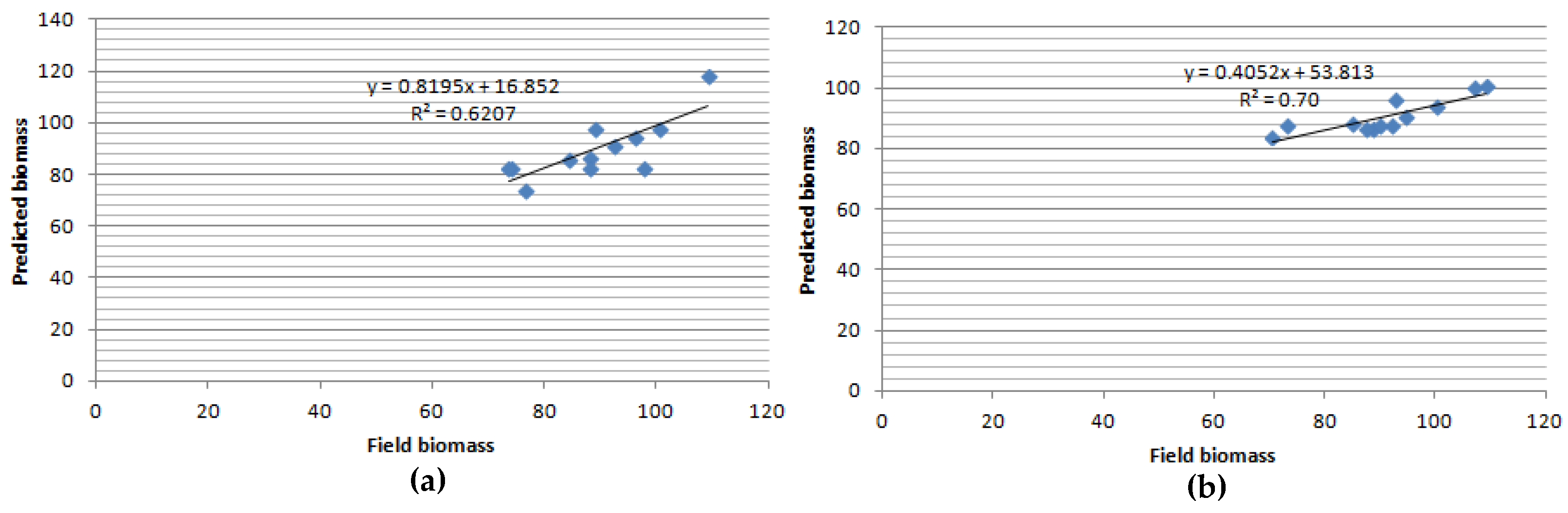
3. Results and Discussion
Above Ground Biomass
4. Conclusions
- (1)
- Due to the lack of access to all areas and high cost and time consuming by ground method, these problems can be overcame by using remote sensing method.
- (2)
- The combination of Sentinel-1 and ALOSPALSAR data illustrated better performance in estimating AGB compared to sentinel-2 data.
- (3)
- GA-RF Model is beneficial and fast to achieve high accuracy in AGB prediction.
- (4)
- The use of feature selection method (GA) to reduce the number of predictor features improved the performance of the RF model.
- (5)
- Effectiveness of texture and decomposition features on AGB calculation.
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGB | Above Ground Biomass |
| GA-RF | Genetic-Random Forest |
| PCA | Principle Component Analysis |
| GIS | Geospatial Information System |
| MPM | Multiplicative Power Model |
| PLSR | Partial Least Square Regression |
| SVR | Support Vector Regression |
| GA-SVM | Genetic-Support Vector Machine |
| DBH | Diameter at Breast Height |
| RTK | Real Time Kinematic |
| ESA | European Space Agency |
| JAXA | Japan Aerospace Exploration Agency |
| SAR | Synthetic Aperture Radar |
| GLCM | Gray Level Co occurrence Matrix |
| VI | Vegetation Index |
| RMSE | Root Mean Square Error |
References
- Luyssaert, S.; Schulze, E.-D.; Börner, A.; Knohl, A.; Hessenmöller, D.; Law, B.E.; Ciais, P.; Grace, J. Old-growth forests as global carbon sinks. Nature 2008, 455, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Lu, D. Aboveground biomass estimation using Landsat TM data in the Brazilian Amazon. Int. J. Remote Sens. 2005, 26, 2509–2525. [Google Scholar] [CrossRef]
- Foody, G.M.; Boyd, D.S.; Cutler, M.E. Predictive relations of tropical forest biomass from Landsat TM data and their transferability between regions. Remote Sens. Environ. 2003, 85, 463–474. [Google Scholar] [CrossRef]
- TSITSI, B. Remote sensing of aboveground forest biomass: A review. Trop. Ecol. 2016, 57, 125–132. [Google Scholar]
- Sinha, S.; Jeganathan, C.; Sharma, L.K.; Nathawat, M.S. A review of radar remote sensing for biomass estimation. Int. J. Environ. Sci. Technol. 2015, 12, 1779–1792. [Google Scholar] [CrossRef]
- Deo, R.K. Modelling and Mapping of Above-Ground Biomass and Carbon Sequestration in the Cool Temperate Forest of North-East China; ITC Enschede: Enschede, The Netherlands, 2008. [Google Scholar]
- Hyde, P.; Nelson, R.; Kimes, D.; Levine, E. Exploring LiDAR–RaDAR synergy—predicting aboveground biomass in a southwestern ponderosa pine forest using LiDAR, SAR and InSAR. Remote Sens. Environ. 2007, 106, 28–38. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Hartig, F.; Latifi, H.; Berger, C.; Hernández, J.; Corvalán, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Ghasemi, N.; Sahebi, M.R.; Mohammadzadeh, A. Biomass estimation of a temperate deciduous forest using wavelet analysis. IEEE Trans. Geosci. Remote Sens. 2012, 51, 765–776. [Google Scholar] [CrossRef]
- Caicoya, A.T.; Kugler, F.; Papathanassiou, K.; Biber, P.; Pretzsch, H. Biomass estimation as a function of vertical forest structure and forest height-Potential and limitations for Radar Remote Sensing. In Proceedings of the 8th European Conference on Synthetic Aperture Radar, Aachen, Germany, 7–10 June 2010; pp. 1–4. [Google Scholar]
- Behera, M.; Tripathi, P.; Mishra, B.; Kumar, S.; Chitale, V.; Behera, S.K. Above-ground biomass and carbon estimates of Shorea robusta and Tectona grandis forests using QuadPOL ALOS PALSAR data. Adv. Space Res. 2016, 57, 552–561. [Google Scholar] [CrossRef]
- West, P.W. Growing Plantation Forests; Springer: Berlin, Germany, 2006. [Google Scholar]
- Pearson, R.L.; Miller, L.D.; Tucker, C.J. Hand-held spectral radiometer to estimate gramineous biomass. Appl. Opt. 1976, 15, 416–418. [Google Scholar] [CrossRef]
- Li, X.; Gar-On Yeh, A.; Wang, S.; Liu, K.; Liu, X.; Qian, J.; Chen, X. Regression and analytical models for estimating mangrove wetland biomass in South China using Radarsat images. Int. J. Remote Sens. 2007, 28, 5567–5582. [Google Scholar] [CrossRef]
- Ghasemi, N.; Sahebi, M.R.; Mohammadzadeh, A. A review on biomass estimation methods using synthetic aperture radar data. Int. J. Geomat. Geosci. 2011, 1, 776–788. [Google Scholar]
- Treuhaft, R.N.; Law, B.E.; Asner, G.P. Forest attributes from radar interferometric structure and its fusion with optical remote sensing. BioScience 2004, 54, 561–571. [Google Scholar] [CrossRef]
- He, Q.; Cao, C.; Chen, E.; Ling, F.; Zhang, H. Relationship between SAR and biomass derived from LiDAR in Mountain areas. In Proceedings of the 2009 2nd Asian-Pacific Conference on Synthetic Aperture Radar, Xi’an, China, 26–30 October 2009; pp. 136–139. [Google Scholar]
- Imhoff, M.L. Radar backscatter and biomass saturation: Ramifications for global biomass inventory. IEEE Trans. Geosci. Remote Sens. 1995, 33, 511–518. [Google Scholar] [CrossRef]
- Le Toan, T.; Beaudoin, A.; Riom, J.; Guyon, D. Relating forest biomass to SAR data. IEEE Trans. Geosci. Remote Sens. 1992, 30, 403–411. [Google Scholar] [CrossRef]
- Cutler, M.; Boyd, D.; Foody, G.; Vetrivel, A. Estimating tropical forest biomass with a combination of SAR image texture and Landsat TM data: An assessment of predictions between regions. ISPRS J. Photogramm. Remote Sens. 2012, 70, 66–77. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using WorldView-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Laurin, G.V.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Del Frate, F.; Guerriero, L.; Pirotti, F.; Valentini, R. Above ground biomass estimation in an African tropical forest with lidar and hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2014, 89, 49–58. [Google Scholar] [CrossRef]
- Karlson, M.; Ostwald, M.; Reese, H.; Sanou, J.; Tankoano, B.; Mattsson, E. Mapping tree canopy cover and aboveground biomass in Sudano-Sahelian woodlands using Landsat 8 and random forest. Remote Sens. 2015, 7, 10017–10041. [Google Scholar] [CrossRef]
- Tavasoli, N.; Arefi, H.; Samiei-Esfahany, S.; Ronoud, Q. Modelling the amount of carbon stock using remote sensing in urban forest and its relationship with land use change. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 1051–1058. [Google Scholar] [CrossRef]
- Nuthammachot, N.; Askar, A.; Stratoulias, D.; Wicaksono, P. Combined use of Sentinel-1 and Sentinel-2 data for improving above-ground biomass estimation. Geocarto Int. 2020, 1–11. [Google Scholar] [CrossRef]
- Brown, S. Estimating Biomass and Biomass Change of Tropical Forests: A Primer; Food & Agriculture Org.: United Nations, 1997; Volume 134. [Google Scholar]
- Brown, S.; Lugo, A.E. Biomass of tropical forests: A new estimate based on forest volumes. Science 1984, 223, 1290–1293. [Google Scholar] [CrossRef]
- Tarmian, A.; Remond, R.; Faezipour, M.; Karimi, A.; Perré, P. Reaction wood drying kinetics: Tension wood in Fagus sylvatica and compression wood in Picea abies. Wood Sci. Technol. 2009, 43, 113–130. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P. Sentinel-2: ESA’s optical high-resolution mission for GMES operational services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Sandwell, D.T.; Myer, D.; Mellors, R.; Shimada, M.; Brooks, B.; Foster, J. Accuracy and resolution of ALOS interferometry: Vector deformation maps of the Father’s Day intrusion at Kilauea. IEEE Trans. Geosci. Remote Sens. 2008, 46, 3524–3534. [Google Scholar] [CrossRef]
- Torres, R.; Snoeij, P.; Geudtner, D.; Bibby, D.; Davidson, M.; Attema, E.; Potin, P.; Rommen, B.; Floury, N.; Brown, M. GMES Sentinel-1 mission. Remote Sens. Environ. 2012, 120, 9–24. [Google Scholar] [CrossRef]
- Ahmed, S.M.; Eldin, F.A.E.; Tarek, A.M. Speckle noise reduction in SAR images using adaptive morphological filter. In Proceedings of the 2010 10th International Conference on Intelligent Systems Design and Applications, Cairo, Egypt, 29 November–1 December 2010; pp. 260–265. [Google Scholar]
- Lee, J.-S. Digital image enhancement and noise filtering by use of local statistics. IEEE Trans. Pattern Anal. Mach. Intell. 1980, 165–168. [Google Scholar] [CrossRef]
- Frost, V.S.; Stiles, J.A.; Shanmugan, K.S.; Holtzman, J.C. A model for radar images and its application to adaptive digital filtering of multiplicative noise. IEEE Trans. Pattern Anal. Mach. Intell. 1982, 157–166. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef]
- Jackson, R.D.; Huete, A.R. Interpreting vegetation indices. Prev. Vet. Med. 1991, 11, 185–200. [Google Scholar] [CrossRef]
- Major, D.; Baret, F.; Guyot, G. A ratio vegetation index adjusted for soil brightness. Int. J. Remote Sens. 1990, 11, 727–740. [Google Scholar] [CrossRef]
- Carlson, T.N.; Ripley, D.A. On the relation between NDVI, fractional vegetation cover, and leaf area index. Remote Sens. Environ. 1997, 62, 241–252. [Google Scholar] [CrossRef]
- Nellis, M.D.; Briggs, J.M. Transformed vegetation index for measuring spatial variation in drought impacted biomass on Konza Prairie, Kansas. Trans. Kansas Acad. Sci. (1903) 1992, 95, 93–99. [Google Scholar] [CrossRef]
- Ashburn, P. The Vegetative Index Number and Crop Identification; Lyndon B. Johnson Space Center: Houston, TX, USA, 1979. [Google Scholar]
- Mohanaiah, P.; Sathyanarayana, P.; GuruKumar, L. Image texture feature extraction using GLCM approach. Int. J. Sci. Res. Publ. 2013, 3, 1–5. [Google Scholar]
- Crist, E.P.; Cicone, R.C. Application of the tasseled cap concept to simulated thematic mapper data. Photogramm. Eng. Remote Sens. 1984, 50, 343–352. [Google Scholar]
- Crist, E.P. A TM tasseled cap equivalent transformation for reflectance factor data. Remote Sens. Environ. 1985, 17, 301–306. [Google Scholar] [CrossRef]
- Richards, J. Thematic mapping from multitemporal image data using the principal components transformation. Remote Sens. Environ. 1984, 16, 35–46. [Google Scholar] [CrossRef]
- Mo, T.; Schmugge, T.J.; Jackson, T.J. Calculations of radar backscattering coefficient of vegetation-covered soils. Remote Sens. Environ. 1984, 15, 119–133. [Google Scholar]
- Lee, J.; Pottier, E. Introduction to the polarimetric target decomposition concept. In Polarimetric Radar Imaging: From Basics to Applications; CRC Press: Boca Raton, FL, USA, 2009; pp. 1–422. [Google Scholar]
- Sun, G.; Ranson, K.J. A three-dimensional radar backscatter model of forest canopies. IEEE Trans. Geosci. Remote Sens. 1995, 33, 372–382. [Google Scholar] [CrossRef]
- Jayasri, P.; Niharika, K.; Joseph, M.; Ryali, H.U.S.; Sarma, C.R.; Kumari, E.S.; Prasad, A. Implementation of RISAT-1 hybrid polarimetric decomposition techniques and analysis using corner reflector data. J. Indian Soc. Remote Sens. 2018, 46, 1005–1012. [Google Scholar] [CrossRef]
- Raney, R.K.; Cahill, J.T.; Patterson, G.W.; Bussey, D.B.J. The m-chi decomposition of hybrid dual-polarimetric radar data with application to lunar craters. J. Geophys. Res. Planets 2012, 117. [Google Scholar] [CrossRef]
- Raney, R.K. M-chi decomposition of imperfect hybrid dual-polarimetric radar data. ESASP 2013, 713, 2. [Google Scholar]
- Raney, R.K. Decomposition of hybrid-polarity SAR data. In PolIn-SAR 2007, Proceedings of the 3rd International Workshop on Science and Applications, Frascati, Italy, 22–26 January 2007; European Space Agency: Noordwijk, The Netherlands, 2007; pp. 22–26. [Google Scholar]
- Cloude, S.R.; Goodenough, D.G.; Chen, H. Compact decomposition theory. IEEE Geosci. Remote Sens. Lett. 2011, 9, 28–32. [Google Scholar] [CrossRef]
- Singh, G.; Venkataraman, G.; Rao, Y. The H/A/Alpha polarimetric decomposition theorem and complex wishart distribution for snow cover monitoring. In Proceedings of the IGARSS 2008—2008 IEEE International Geoscience and Remote Sensing Symposium, Boston, MA, USA, 7–11 July 2008; pp. IV-1081–IV-1084. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Mitchell, M. An Introduction to Genetic Algorithms; MIT Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Ali, E.E.E.; Elamin, E. A Proposed Genetic Algorithm Selection Method; King Saud University, College of Computer and Information Sciences: Riyadh, Saudi Arabia, 2006. [Google Scholar]
- Oh, I.-S.; Lee, J.-S.; Moon, B.-R. Hybrid genetic algorithms for feature selection. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 1424–1437. [Google Scholar]
- Bernal, B.; Murray, L.T.; Pearson, T.R. Global carbon dioxide removal rates from forest landscape restoration activities. Carbon Balance Manag. 2018, 13, 22. [Google Scholar] [CrossRef]


| Vegetation Vegetation Indexdex | EquationEquation |
|---|---|
| Ratio vegetation index | NIR/R |
| Normalized difference Vegetation index (NDVI) | (NIR-R)/(NIR+R) |
| Transformed vegetation index (TVI) | |
| Ashburn vegetation index (AVI) | 2.0[800:1100]-[600:700] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavasoli, N.; Arefi, H. Comparison of Capability of SAR and Optical Data in Mapping Forest above Ground Biomass Based on Machine Learning. Environ. Sci. Proc. 2021, 5, 13. https://doi.org/10.3390/IECG2020-07916
Tavasoli N, Arefi H. Comparison of Capability of SAR and Optical Data in Mapping Forest above Ground Biomass Based on Machine Learning. Environmental Sciences Proceedings. 2021; 5(1):13. https://doi.org/10.3390/IECG2020-07916
Chicago/Turabian StyleTavasoli, Negar, and Hossein Arefi. 2021. "Comparison of Capability of SAR and Optical Data in Mapping Forest above Ground Biomass Based on Machine Learning" Environmental Sciences Proceedings 5, no. 1: 13. https://doi.org/10.3390/IECG2020-07916
APA StyleTavasoli, N., & Arefi, H. (2021). Comparison of Capability of SAR and Optical Data in Mapping Forest above Ground Biomass Based on Machine Learning. Environmental Sciences Proceedings, 5(1), 13. https://doi.org/10.3390/IECG2020-07916






