Phenotypic Evaluation of Recombinant Inbred Lines for Sodicity Tolerance at Reproductive Stage in Rice †
Abstract
1. Introduction
2. Materials and Methods
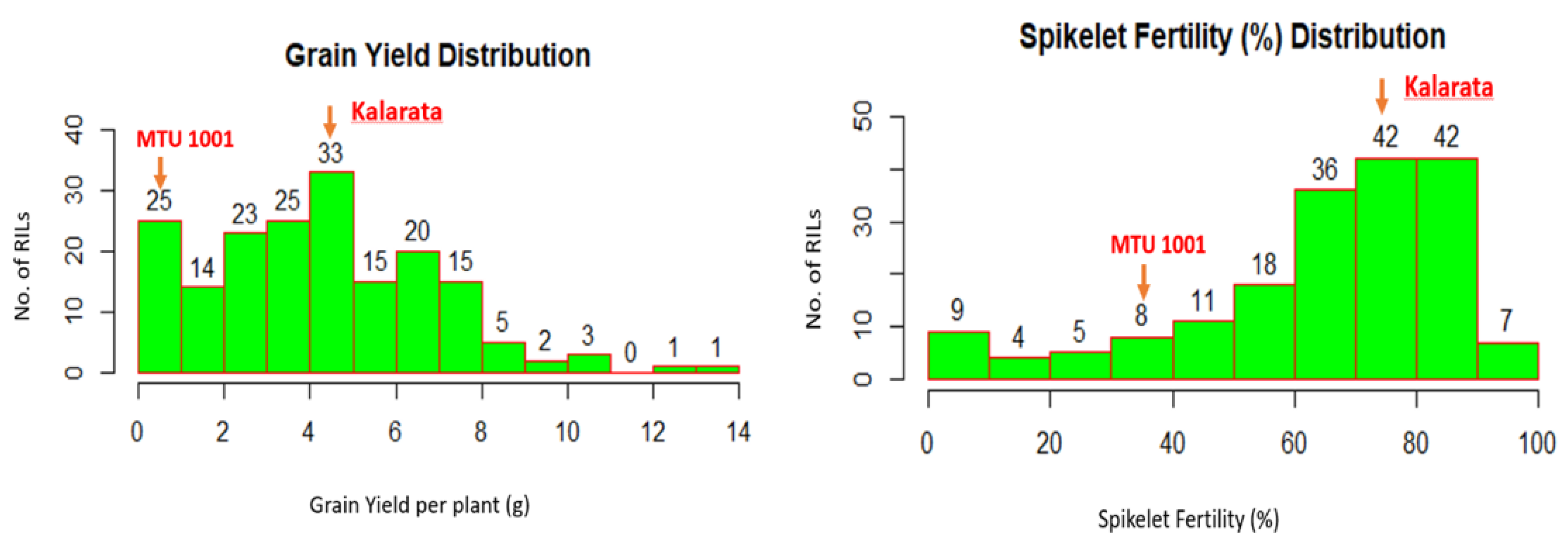
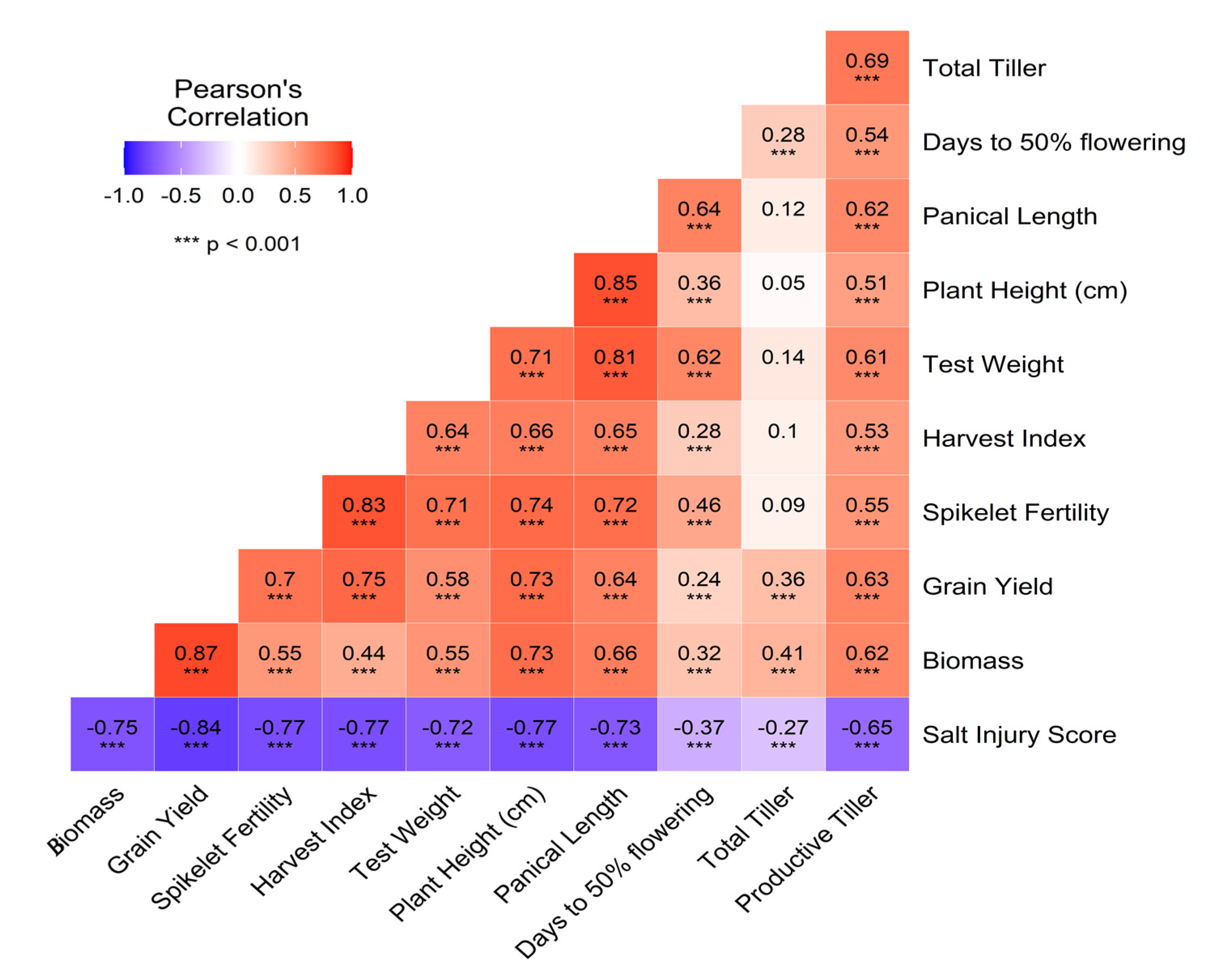
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- IIRR, 2015; Vision 2050. ICAR-Indian Institute of Rice Research: Hyderabad, India, 2015; p. 2.
- Hoang, T.; Tran, T.; Nguyen, T.; Williams, B.; Wurm, P.; Bellairs, S.; Mundree, S. Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agronomy 2016, 6, 54. [Google Scholar] [CrossRef]
- FAO and ITPS. Status of the World’s Soil Resources (SWSR)—Main Report; Food and Agriculture Organization of the United Nations and Intergovernmental Technical Panel on Soils: Rome, Italy, 2015.
- Morales, S.G.; Trejo-Téllez, L.I.; Gómez Merino, F.C.; Caldana, C.; Espinosa-Victoria, D.; Herrera Cabrera, B.E. Growth, photosynthetic activity, and potassium and sodium concentration in rice plants under salt stress. Acta Scientiarum. Agron. 2012, 34, 317–324. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Gregorio, G.; Subudhi, P.K. Genetic variation in southern USA rice genotypes for seedling salinity tolerance. Front. Plant Sci. 2015, 6, 374. [Google Scholar] [CrossRef]
- Krishnamurthy, S.L.; Gautam, R.K.; Sharma, P.C.; Sharma, D.K. Effect of different salt stresses on agro-morphological traits and utilisation of salt stress indices for reproductive stage salt tolerance in Rice. Field Crops Res. 2016, 190, 26–33. [Google Scholar] [CrossRef]
- Kranto, S.; Chankaew, S.; Monkham, T. Theerakulpisut, P.; Sanitchon, J. Evaluation for salt tolerance in rice using multiple screening methods. J. Agric. Sci. Technol. 2016, 18, 1921–1931. [Google Scholar]
- IRRI (2014); Standard Evaluation System for Rice. 5th ed. International Rice Research Institute: Los Banos, The Philippines.
- Krishnamurthy, S.L.; Sharma, S.K.; Gautam, R.K.; Kumar, V. Path and association analysis and stress indices for salinity tolerance traits in promising rice (Oryza sativa L.) Genotypes. Cereal Res. Commun. 2014, 42, 474–483. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Z.; Chen, Y.; Yang, C.; Shi, D. Effects of salt and alkali stresses on growth and ion balance in rice (Oryza sativa L.). Plant Soil Environ. 2011, 57, 286–294. [Google Scholar] [CrossRef]
- Chakraborty, S.; Das, P.K.; Guha, B.; Sarmah, K.K.; Barman, B. Quantitative genetic analysis for yield and yield components in Boro Rice (Oryza sativa L.). Not. Sci. Biol. 2010, 2, 117–120. [Google Scholar] [CrossRef]
- Sanghera, G.S.; Kashyap, S.C. Genetic parameters and selection indices in F3 progenies of hill rice genotypes. Not. Sci. Biol. 2012, 4, 110–114. [Google Scholar] [CrossRef][Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nain, A.; Krishnamurthy, S.L.; Sharma, P.C.; Lokeshkumar, B.M.; Kumar, M.; Warraich, A.S. Phenotypic Evaluation of Recombinant Inbred Lines for Sodicity Tolerance at Reproductive Stage in Rice. Environ. Sci. Proc. 2022, 16, 47. https://doi.org/10.3390/environsciproc2022016047
Nain A, Krishnamurthy SL, Sharma PC, Lokeshkumar BM, Kumar M, Warraich AS. Phenotypic Evaluation of Recombinant Inbred Lines for Sodicity Tolerance at Reproductive Stage in Rice. Environmental Sciences Proceedings. 2022; 16(1):47. https://doi.org/10.3390/environsciproc2022016047
Chicago/Turabian StyleNain, Ashish, Saraswathipura L. Krishnamurthy, Parbodh C. Sharma, Bayragondlu M. Lokeshkumar, Mukesh Kumar, and Arvinder S. Warraich. 2022. "Phenotypic Evaluation of Recombinant Inbred Lines for Sodicity Tolerance at Reproductive Stage in Rice" Environmental Sciences Proceedings 16, no. 1: 47. https://doi.org/10.3390/environsciproc2022016047
APA StyleNain, A., Krishnamurthy, S. L., Sharma, P. C., Lokeshkumar, B. M., Kumar, M., & Warraich, A. S. (2022). Phenotypic Evaluation of Recombinant Inbred Lines for Sodicity Tolerance at Reproductive Stage in Rice. Environmental Sciences Proceedings, 16(1), 47. https://doi.org/10.3390/environsciproc2022016047





