Toxicogenomics of Arsenic, Lead and Mercury: The Toxic Triad
Abstract
1. Introduction
2. Materials and Methods
2.1. Target Elements
2.2. Metal–Gene Interactions
2.3. Analyses of Gene–Metal Interactions
2.4. Gene Ontology, Enriched Diseases, Enriched Pathways, and Gene–Gene Interaction Network
2.5. Genotoxic and Carcinogenic Activity
3. Results
3.1. Metal–Gene Interactions
3.2. Profile of Metal–Gene Interactions
3.3. Gene Ontology, Enriched Diseases and Enriched Pathways
3.4. Genotoxic and Carcinogenic Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murguía, D.I.; Bringezu, S.; Schaldach, R. Global direct pressures on biodiversity by large-scale metal mining: Spatial distribution and implications for conservation. J. Environ. Manag. 2016, 180, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Luckeneder, S.; Giljum, S.; Schaffartzik, A.; Maus, V.; Tost, M. Surge in global metal mining threatens vulnerable ecosystems. Glob. Environ. Change 2021, 69, 102303. [Google Scholar] [CrossRef]
- Maus, V.; Giljum, S.; da Silva, D.M.; Gutschlhofer, J.; da Rosa, R.P.; Luckeneder, S.; Gass, S.L.B.; Lieber, M.; McCallum, I. An update on global mining land use. Sci. Data 2022, 9, 433. [Google Scholar] [CrossRef]
- Mosquera-Guerra, F.; Trujillo, F.; Parks, D.; Oliveira-da-Costa, M.; Van Damme, P.A.; Echeverría, A.; Franco, N.; Carva-jal-Castro, J.D.; Mantilla-Meluk, H.; Marmontel, M.; et al. Mercury in Populations of River Dolphins of the Amazon and Orinoco Basins. Ecohealth 2019, 16, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Ellwanger, J.H.; Chies, J.A.B. Brazil’s heavy metal pollution harms humans and ecosystems. Sci. One Health 2023, 2, 100019. [Google Scholar] [CrossRef]
- Fritz, B.; Peregovich, B.; da Silva Tenório, L.; da Silva Alves, A.C.; Schmidt, M. Mercury and CO2 emissions from artisanal gold mining in Brazilian Amazon rainforest. Nat. Sustain. 2024, 7, 15–22. [Google Scholar] [CrossRef]
- Quan, S.X.; Yan, B.; Yang, F.; Li, N.; Xiao, X.M.; Fu, J.M. Spatial distribution of heavy metal contamination in soils near a primitive e-waste recycling site. Environ. Sci. Pollut. Res. Int. 2015, 22, 1290–1298. [Google Scholar] [CrossRef]
- Awasthi, A.K.; Zeng, X.; Li, J. Environmental pollution of electronic waste recycling in India: A critical review. Environ. Pollut. 2016, 211, 259–270. [Google Scholar] [CrossRef]
- Sundseth, K.; Pacyna, J.M.; Pacyna, E.G.; Pirrone, N.; Thorne, R.J. Global Sources and Pathways of Mercury in the Context of Human Health. Int. J. Environ. Res. Public Health 2017, 14, 105. [Google Scholar] [CrossRef]
- Quinteros, E.; Ribó, A.; Mejía, R.; López, A.; Belteton, W.; Comandari, A.; Orantes, C.M.; Pleites, E.B.; Hernández, C.E.; López, D.L. Heavy metals and pesticide exposure from agricultural activities and former agrochemical factory in a Salvadoran rural community. Environ. Sci. Pollut. Res. Int. 2017, 24, 1662–1676. [Google Scholar] [CrossRef]
- Reboredo, F.; Simões, M.; Jorge, C.; Mancuso, M.; Martinez, J.; Guerra, M.; Ramalho, J.C.; Pessoa, M.F.; Lidon, F. Metal content in edible crops and agricultural soils due to intensive use of fertilizers and pesticides in Terras da Costa de Caparica (Portugal). Environ. Sci. Pollut. Res. Int. 2019, 26, 2512–2522. [Google Scholar] [CrossRef]
- Mamtani, R.; Stern, P.; Dawood, I.; Cheema, S. Metals and disease: A global primary health care perspective. J. Toxicol. 2011, 2011, 319136. [Google Scholar] [CrossRef] [PubMed]
- Marti, D.; Hanrahan, D.; Sanchez-Triana, E.; Wells, M.; Corra, L.; Hu, H.; Breysse, P.N.; Laborde, A.; Caravanos, J.; Bertollini, R.; et al. Structured expert judgement approach of the health impact of various chemicals and classes of chemicals. PLoS ONE 2024, 19, e0298504. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; Costa, M. Arsenic: A Global Environmental Challenge. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Lauria, G.; Catalano, A.; Carocci, A.; Sinicropi, M.S. Arsenic: A Review on a Great Health Issue Worldwide. Appl. Sci. 2022, 12, 6184. [Google Scholar] [CrossRef]
- Wani, A.L.; Ara, A.; Usmani, J.A. Lead toxicity: A review. Interdiscip. Toxicol. 2015, 8, 55–64. [Google Scholar] [CrossRef]
- Raj, K.; Das, A.P. Lead pollution: Impact on environment and human health and approach for a sustainable solution. Environ. Chem. Ecotox. 2023, 5, 79–85. [Google Scholar] [CrossRef]
- Fuller, R.; Porterfield, K.; Hanrahan, D.; Hu, H. Cumulative population blood lead levels. BMJ Glob. Health 2025, 10, e018145. [Google Scholar] [CrossRef]
- Patra, M.; Bhowmik, N.; Bandopadhyay, B.; Sharma, A. Comparison of mercury, lead and arsenic with respect to genotoxic effects on plant systems and the development of genetic tolerance. Environ. Exp. Bot. 2004, 52, 199–223. [Google Scholar] [CrossRef]
- Musah, B.I. Effects of heavy metals and metalloids on plant-animal interaction and biodiversity of terrestrial ecosystems-an overview. Environ. Monit. Assess. 2025, 197, 12. [Google Scholar] [CrossRef]
- Hou, D.; Jia, X.; Wang, L.; McGrath, S.P.; Zhu, Y.G.; Hu, Q.; Zhao, F.J.; Bank, M.S.; O’Connor, D.; Nriagu, J. Global soil pollution by toxic metals threatens agriculture and human health. Science 2025, 388, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Billionnet, C.; Sherrill, D.; Annesi-Maesano, I.; GERIE Study. Estimating the health effects of exposure to multi-pollutant mixture. Ann. Epidemiol. 2012, 22, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cobbina, S.J.; Mao, G.; Xu, H.; Zhang, Z.; Yang, L. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ. Sci. Pollut. Res. Int. 2016, 23, 8244–8259. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, B.O.; Ezejiofor, A.N.; Igweze, Z.N.; Orisakwe, O.E. Heavy Metal Mixture Exposure and Effects in Developing Nations: An Update. Toxics 2018, 6, 65. [Google Scholar] [CrossRef]
- Davis, A.P.; Murphy, C.G.; Rosenstein, M.C.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database facilitates identification and understanding of chemical-gene-disease associations: Arsenic as a case study. BMC Med. Genomics 2008, 1, 48. [Google Scholar] [CrossRef]
- Koedrith, P.; Kim, H.L.; Seo, Y.R. Integrative toxicogenomics-based approach to risk assessment of heavy metal mix-tures/complexes: Strategies and challenges. Mol. Cell. Toxicol. 2015, 11, 265–276. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Duraikannan, V.; Perumal, E. Toxicogenomic analysis of physiologically important metals: An integrated in silico approach. Food Chem. Toxicol. 2023, 178, 113895. [Google Scholar] [CrossRef]
- Meier, M.J.; Harrill, J.; Johnson, K.; Thomas, R.S.; Tong, W.; Rager, J.E.; Yauk, C.L. Progress in toxicogenomics to protect human health. Nat. Rev. Genet. 2025, 26, 105–122. [Google Scholar] [CrossRef]
- Chepelev, N.L.; Moffat, I.D.; Labib, S.; Bourdon-Lacombe, J.; Kuo, B.; Buick, J.K.; Lemieux, F.; Malik, A.I.; Halappanavar, S.; Williams, A.; et al. Integrating toxicogenomics into human health risk assessment: Lessons learned from the benzo[a]pyrene case study. Crit. Rev. Toxicol. 2015, 45, 44–52. [Google Scholar] [CrossRef]
- Schmitz-Spanke, S. Toxicogenomics—What Added Value Do These Approaches Provide for Carcinogen Risk Assessment? Environ. Res. 2019, 173, 157–164. [Google Scholar] [CrossRef]
- Royal Society of Chemistry. Periodic Table. 2025. Available online: https://periodic-table.rsc.org/ (accessed on 23 March 2025).
- World Health Organization. 10 Chemicals of Public Health Concern. 2020. Available online: https://www.who.int/news-room/photo-story/detail/10-chemicals-of-public-health-concern (accessed on 23 March 2025).
- Pure Earth; Green Cross Switzerland. World’s Worst Pollution Problems 2015: The New Top Six Toxic Threats: A Priority List for Remediation; Pure Earth: New York, NY, USA, 2015. [Google Scholar]
- Pure Earth; Green Cross Switzerland. World’s Worst Pollution Problems 2016: The Toxics Beneath Our Feet; Pure Earth: New York, NY, USA, 2016. [Google Scholar]
- CTD—The Comparative Toxicogenomics Database. 2025. Available online: https://ctdbase.org/ (accessed on 23 March 2025).
- Davis, A.P.; Wiegers, T.C.; Sciaky, D.; Barkalow, F.; Strong, M.; Wyatt, B.; Wiegers, J.; McMorran, R.; Abrar, S.; Mattingly, C.J. Comparative Toxicogenomics Database’s 20th Anniversary: Update 2025. Nucleic Acids Res. 2025, 53, D1328–D1334. [Google Scholar] [CrossRef] [PubMed]
- Ellwanger, J.H.; Chies, J.A.B. Toxicogenomics of the C-C chemokine receptor type 5 (CCR5): Exploring the potential impacts of chemical-CCR5 interactions on inflammation and human health. Food Chem. Toxicol. 2024, 186, 114511. [Google Scholar] [CrossRef]
- Ziliotto, M.; Chies, J.A.B.; Ellwanger, J.H. Toxicogenomics of persistent organic pollutants: Potential impacts on biodiversity and infectious diseases. Anthropocene 2024, 48, 100450. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Ziliotto, M.; Chies, J.A.B. Toxicogenomics of glutathione S-transferase (GST) gene family members: Chemical-gene interactions and potential implications of gene deletions. Comput. Biol. Med. 2025, 189, 110025. [Google Scholar] [CrossRef]
- CTD—The Comparative Toxicogenomics Database. VennViewer. 2025. Available online: https://ctdbase.org/tools/vennViewer.go (accessed on 24 March 2025).
- CTD—The Comparative Toxicogenomics Database. Set Analyzer. 2025. Available online: https://ctdbase.org/tools/analyzer.go (accessed on 29 March 2025).
- CTD—The Comparative Toxicogenomics Database. Help: Set Analyzer. 2025. Available online: https://ctdbase.org/help/analyzerHelp.jsp (accessed on 30 March 2025).
- Oughtred, R.; Breitkreutz, B.J.; Boucher, L.; Chang, C.S.; Rust, J.M.; Chatraryamontri, A.; Kolas, N.; O’Donnell, L.; Theesfeld, C.L.; Stark, C.; et al. The BioGRID interaction database: Integration of genetic, protein and chemical interactions and an improved network viewer. Protein Sci. 2016, 5, 745. [Google Scholar] [CrossRef]
- BioGRID BioGRID 4.4. Available online: https://thebiogrid.org/ (accessed on 22 April 2025).
- Williams, A.J.; Grulke, C.M.; Edwards, J.; McEachran, A.D.; Mansouri, K.; Baker, N.C.; Patlewicz, G.; Shah, I.; Wambaugh, J.F.; Judson, R.S.; et al. The CompTox Chemistry Dashboard: A community data resource for environmental chemistry. J. Cheminform. 2017, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- EPA—US Environmental Protection Agency. CompTox Chemicals Dashboard v2.5.2. Available online: https://comptox.epa.gov/dashboard/ (accessed on 24 March 2025).
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic Mechanisms of Five Heavy Metals: Mercury, Lead, Chromium, Cadmium, and Arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.N.; Tangpong, J.; Rahman, M. Toxicodynamics of Lead, Cadmium, Mercury and Arsenic-induced kidney toxicity and treatment strategy: A mini review. Toxicol. Rep. 2018, 5, 704–713. [Google Scholar] [CrossRef]
- Chang, C.H.; Liu, C.S.; Liu, H.J.; Huang, C.P.; Huang, C.Y.; Hsu, H.T.; Liou, S.H.; Chung, C.J. Association between levels of urinary heavy metals and increased risk of urothelial carcinoma. Int. J. Urol. 2016, 23, 233–239. [Google Scholar] [CrossRef]
- Simeonova, P.P.; Wang, S.; Toriuma, W.; Kommineni, V.; Matheson, J.; Unimye, N.; Kayama, F.; Harki, D.; Ding, M.; Vallyathan, V.; et al. Arsenic mediates cell proliferation and gene expression in the bladder epithelium: Association with activating protein-1 transactivation. Cancer Res. 2000, 60, 3445–3453. [Google Scholar]
- El-Agrody, E.; Abol-Enein, H.; Mortada, W.I.; Awadalla, A.; Tarabay, H.H.; Elkhawaga, O.A. Does the Presence of Heavy Metals Influence the Gene Expression and Oxidative Stress in Bladder Cancer? Biol. Trace Elem. Res. 2024, 202, 3475–3482. [Google Scholar] [CrossRef] [PubMed]
- García-Lestón, J.; Méndez, J.; Pásaro, E.; Laffon, B. Genotoxic effects of lead: An updated review. Environ. Int. 2010, 36, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Xi, S. A review on arsenic carcinogenesis: Epidemiology, metabolism, genotoxicity and epigenetic changes. Regul. Toxicol. Pharmacol. 2018, 99, 78–88. [Google Scholar] [CrossRef]
- Nagaraju, R.; Kalahasthi, R.; Balachandar, R.; Bagepally, B.S. Association between lead exposure and DNA damage (genotoxicity): Systematic review and meta-analysis. Arch. Toxicol. 2022, 96, 2899–2911. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Joardar, M.; Chowdhury, N.R.; De, A.; Mridha, D.; Roychowdhury, T. Arsenic toxicity in livestock growing in arsenic endemic and control sites of West Bengal: Risk for human and environment. Environ. Geochem. Health 2021, 43, 3005–3025. [Google Scholar] [CrossRef]
- Bundschuh, J.; Niazi, N.K.; Alam, M.A.; Berg, M.; Herath, I.; Tomaszewska, B.; Maity, J.P.; Ok, Y.S. Global arsenic dilemma and sustainability. J. Hazard Mater. 2022, 436, 129197. [Google Scholar] [CrossRef]
- Issanov, A.; Adewusi, B.; Saint-Jacques, N.; Dummer, T.J.B. Arsenic in drinking water and lung cancer: A systematic review of 35 years of evidence. Toxicol. Appl. Pharmacol. 2024, 483, 116808. [Google Scholar] [CrossRef]
- Ozturk, M.; Metin, M.; Altay, V.; Bhat, R.A.; Ejaz, M.; Gul, A.; Unal, B.T.; Hasanuzzaman, M.; Nibir, L.; Nahar, K.; et al. Arsenic and Human Health: Genotoxicity, Epigenomic Effects, and Cancer Signaling. Biol. Trace Elem. Res. 2022, 200, 988–1001. [Google Scholar] [CrossRef]
- Chen, T.; Dai, K.; Wu, H. Effect of lead exposure on respiratory health: A systematic review and meta-analysis. Air Qual. Atmos. Health 2024, 17, 3031–3044. [Google Scholar] [CrossRef]
- Hydeskov, H.B.; Arnemo, J.M.; Lloyd Mills, C.; Gentle, L.K.; Uzal, A. A Global Systematic Review of Lead (Pb) Exposure and its Health Effects in Wild Mammals. J. Wildl. Dis. 2024, 60, 285–297. [Google Scholar] [CrossRef]
- Bhowmik, N.; Patra, M. Assessment of genotoxicity of inorganic mercury in rats in vivo using both chromosomal aberration and comet assays. Toxicol. Ind. Health 2015, 31, 588–594. [Google Scholar] [CrossRef]
- Pastor-Sierra, K.; Espitia-Pérez, L.; Espitia-Pérez, P.; Peñata-Taborda, A.; Brango, H.; Galeano-Páez, C.; Bru-Cordero, O.E.; Palma-Parra, M.; Díaz, S.M.; Trillos, C.; et al. Micronuclei frequency and exposure to chemical mixtures in three Colombian mining populations. Sci. Total. Environ. 2023, 901, 165789. [Google Scholar] [CrossRef] [PubMed]
- Skalny, A.V.; Aschner, M.; Sekacheva, M.I.; Santamaria, A.; Barbosa, F.; Ferrer, B.; Aaseth, J.; Paoliello, M.M.B.; Rocha, J.B.T.; Tinkov, A.A. Mercury and cancer: Where are we now after two decades of research? Food Chem. Toxicol. 2022, 164, 113001. [Google Scholar] [CrossRef]
- Crespo-López, M.E.; Macêdo, G.L.; Pereira, S.I.; Arrifano, G.P.; Picanço-Diniz, D.L.; do Nascimento, J.L.; Herculano, A.M. Mercury and human genotoxicity: Critical considerations and possible molecular mechanisms. Pharmacol. Res. 2009, 60, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Smedley, P.L.; Kinniburgh, D.G. A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 2002, 17, 517–568. [Google Scholar] [CrossRef]
- Izadi, L.N.; Tamadoni, A.; Siebecker, M.G.; Sricharoenvech, P.; Barreto, M.S.C.; Fischel, M.H.H.; Tappero, R.; Sparks, D.L. Hurricanes and turbulent floods threaten arsenic-contaminated coastal soils and vulnerable communities. Environ. Int. 2025, 200, 109479. [Google Scholar] [CrossRef]
- Clarkson, T.W.; Magos, L. The toxicology of mercury and its chemical compounds. Crit. Rev. Toxicol. 2008, 36, 609–662. [Google Scholar] [CrossRef]
- Consoli, V.; Sorrenti, V.; Grosso, S.; Vanella, L. Heme Oxygenase-1 Signaling and Redox Homeostasis in Physiopathological Conditions. Biomolecules 2021, 11, 589. [Google Scholar] [CrossRef]
- Anwar, S.; Alrumaihi, F.; Sarwar, T.; Babiker, A.Y.; Khan, A.A.; Prabhu, S.V.; Rahmani, A.H. Exploring Therapeutic Potential of Catalase: Strategies in Disease Prevention and Management. Biomolecules 2024, 14, 697. [Google Scholar] [CrossRef]
- Preethi, S.; Arthiga, K.; Patil, A.B.; Spandana, A.; Jain, V. Review on NAD(P)H dehydrogenase quinone 1 (NQO1) pathway. Mol. Biol. Rep. 2022, 49, 8907–8924. [Google Scholar] [CrossRef]
- Murakami, S.; Kusano, Y.; Okazaki, K.; Akaike, T.; Motohashi, H. NRF2 signalling in cytoprotection and metabolism. Br. J. Pharmacol. 2023; in press. [Google Scholar] [CrossRef] [PubMed]
- Fara, A.; Mitrev, Z.; Rosalia, R.A.; Assas, B.M. Cytokine storm and COVID-19: A chronicle of pro-inflammatory cytokines. Open Biol. 2020, 10, 200160. [Google Scholar] [CrossRef]
- Kumar, S. Caspase function in programmed cell death. Cell Death Differ. 2007, 14, 32–43. [Google Scholar] [CrossRef]
- NCBI—National Center for Biotechnology Information, National Library of Medicine. Gene. Available online: https://www.ncbi.nlm.nih.gov/gene/?term= (accessed on 10 June 2025).
- Skalny, A.V.; Lima, T.R.R.; Ke, T.; Zhou, J.C.; Bornhorst, J.; Alekseenko, S.I.; Aaseth, J.; Anesti, O.; Sarigiannis, D.A.; Tsatsakis, A.; et al. Toxic metal exposure as a possible risk factor for COVID-19 and other respiratory infectious diseases. Food Chem. Toxicol. 2020, 146, 111809. [Google Scholar] [CrossRef] [PubMed]
- Mahon, M.B.; Sack, A.; Aleuy, O.A.; Barbera, C.; Brown, E.; Buelow, H.; Civitello, D.J.; Cohen, J.M.; de Wit, L.A.; Forstchen, M.; et al. A meta-analysis on global change drivers and the risk of infectious disease. Nature 2024, 629, 830–836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, J.; Zhang, K.; Shi, J.; Gao, Y.; Zheng, J.; He, J.; Zhang, J.; Song, Y.; Zhang, R.; et al. Association between heavy metals exposure and persistent infections: The mediating role of immune function. Front. Public Health 2024, 12, 1367644. [Google Scholar] [CrossRef]
- Mattes, W.B. Cross-species comparative toxicogenomics as an aid to safety assessment. Expert Opin. Drug Metab. Toxicol. 2006, 2, 859–874. [Google Scholar] [CrossRef]
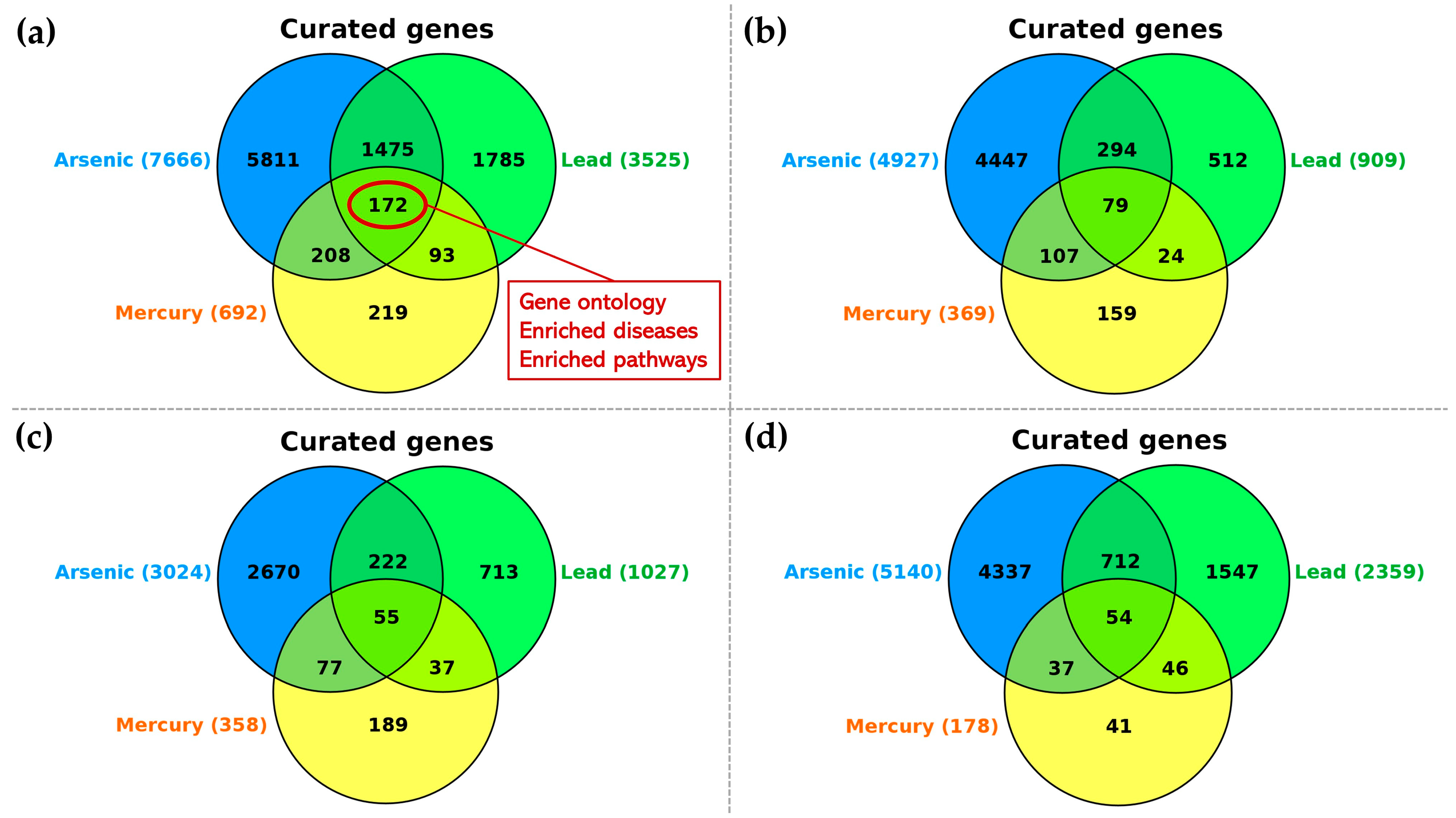


| Arsenic (n = 7666 Interacting Genes) | Lead (n = 3525 Interacting Genes) | Mercury (n = 692 Interacting Genes) | ||||||
|---|---|---|---|---|---|---|---|---|
| Top-20 Gene Names | Number of Metal–Gene Interactions | Organism Number | Top-20 Gene Names | Number of Metal–Gene Interactions | Organism Number | Top-20 Gene Names | Number of Metal–Gene Interactions | Organism Number |
| CXCL8 | 101 | 1 | TNF | 61 | 4 | CYP1A1 | 41 | 2 |
| CAT | 87 | 8 | CYP1A1 | 57 | 3 | HMOX1 | 37 | 4 |
| NFE2L2 | 77 | 4 | CAT | 56 | 7 | NQO1 | 25 | 2 |
| CASP3 | 68 | 5 | MT1 | 54 | 1 | TNF | 21 | 4 |
| AS3MT | 67 | 5 | HMOX1 | 53 | 5 | IL6 | 19 | 4 |
| HMOX1 | 62 | 4 | ALAD | 49 | 4 | ABCC2 | 17 | 5 |
| MAPK1 | 62 | 6 | CASP3 | 49 | 4 | NFE2L2 | 16 | 4 |
| MAPK3 | 59 | 6 | MT2 | 48 | 2 | MT2 | 15 | 6 |
| TNF | 53 | 3 | NFE2L2 | 42 | 4 | MT1 | 14 | 5 |
| GSR | 42 | 8 | APP | 41 | 3 | CAT | 13 | 4 |
| VIM | 42 | 2 | NQO1 | 37 | 4 | GSTP1 | 13 | 3 |
| CDH1 | 39 | 1 | PTGS2 | 36 | 4 | IFNG | 13 | 2 |
| APOE | 38 | 2 | IL6 | 28 | 4 | GSTA1 | 12 | 2 |
| ERBB2 | 37 | 1 | IL1B | 26 | 5 | ALB | 11 | 3 |
| NQO1 | 36 | 4 | SOD1 | 26 | 9 | MT3 | 11 | 3 |
| TP53 | 36 | 6 | HSPA5 | 25 | 4 | CASP3 | 9 | 2 |
| IL6 | 35 | 3 | BCL2 | 24 | 4 | CRYZ | 9 | 1 |
| SNAI1 | 35 | 1 | MAPK3 | 24 | 3 | GSTA2 | 9 | 3 |
| SQSTM1 | 31 | 4 | BAX | 23 | 4 | RELA | 8 | 4 |
| ATF3 | 31 | 2 | RELA | 22 | 4 | SOD1, MT1A, MT2A * | 8 | 3 |
| Biological Processes | Molecular Functions | Cellular Components | |||
|---|---|---|---|---|---|
| Gene Ontology Term | Corrected p-Value | Gene Ontology Term | Corrected p-Value | Gene Ontology Term | Corrected p-Value |
| cellular response to chemical stimulus | 1.14 × 10−78 | Binding | 3.04 × 10−53 | cytoplasm | 7.07 × 10−48 |
| response to chemical | 1.28 × 10−74 | protein binding | 1.69 × 10−52 | cellular anatomical entity | 7.91 × 10−47 |
| response to stress | 1.28 × 10−70 | identical protein binding | 6.64 × 10−41 | intracellular anatomical structure | 2.45 × 10−39 |
| response to stimulus | 1.16 × 10−68 | enzyme binding | 1.41 × 10−29 | membrane-bounded organelle | 6.35 × 10−37 |
| response to organic substance | 2.67 × 10−62 | catalytic activity | 4.73 × 10−21 | intracellular membrane-bounded organelle | 2.30 × 10−36 |
| response to external stimulus | 1.14 × 10−59 | heterocyclic compound binding | 3.14 × 10−20 | cytosol | 2.35 × 10−34 |
| cellular response to stimulus | 2.09 × 10−59 | organic cyclic compound binding | 7.01 × 10−20 | organelle | 1.80 × 10−33 |
| positive regulation of biological process | 2.89 × 10−59 | molecular function regulator activity | 2.16 × 10−19 | intracellular organelle | 3.84 × 10−33 |
| negative regulation of biological process | 7.67 × 10−56 | antioxidant activity | 1.58 × 10−18 | extracellular region | 3.77 × 10−29 |
| biological regulation | 1.95 × 10−55 | protein dimerization activity | 3.77 × 10−18 | endomembrane system | 4.78 × 10−27 |
| regulation of biological process | 3.15 × 10−55 | protein-containing complex binding | 6.43 × 10−17 | intracellular organelle lumen | 9.04 × 10−26 |
| response to oxygen-containing compound | 5.97 × 10−55 | ion binding | 9.93 × 10−17 | membrane-enclosed lumen | 9.04 × 10−26 |
| positive regulation of cellular process | 1.43 × 10−53 | protein homodimerization activity | 3.96 × 10−15 | organelle lumen | 9.04 × 10−26 |
| negative regulation of cellular process | 2.84 × 10−53 | oxidoreductase activity | 6.02 × 10−15 | extracellular space | 1.22 × 10−25 |
| cellular process | 4.04 × 10−53 | receptor ligand activity | 2.42 × 10−14 | cell periphery | 2.19 × 10−24 |
| metabolic process | 1.79 × 10−50 | signaling receptor activator activity | 3.52 × 10−14 | plasma membrane | 5.20 × 10−22 |
| cellular metabolic process | 8.36 × 10−50 | molecular function activator activity | 6.47 × 10−14 | membrane | 1.14 × 10−20 |
| apoptotic process | 1.03 × 10−49 | signaling receptor regulator activity | 1.13 × 10−13 | nucleus | 2.22 × 10−19 |
| cellular response to organic substance | 1.64 × 10−49 | signaling receptor binding | 1.89 × 10−13 | vesicle | 1.02 × 10−18 |
| programmed cell death | 1.47 × 10−48 | ubiquitin protein ligase binding | 9.62 × 10−12 | cytoplasmic vesicle | 1.43 × 10−16 |
| Disease | Corrected p-Value |
|---|---|
| Pathologic processes | 5.72 × 10−102 |
| Pathological conditions, signs and symptoms | 1.15 × 10−97 |
| Digestive system diseases | 1.23 × 10−94 |
| Liver diseases | 9.37 × 10−92 |
| Cardiovascular diseases | 1.86 × 10−87 |
| Urologic diseases | 1.22 × 10−81 |
| Neoplasms by site | 2.00 × 10−81 |
| Vascular diseases | 3.42 × 10−81 |
| Chemically-induced disorders | 1.72 × 10−79 |
| Neoplasms | 4.76 × 10−79 |
| Kidney diseases | 3.91 × 10−77 |
| Male urogenital diseases | 7.47 × 10−77 |
| Nutritional and metabolic diseases | 4.56 × 10−76 |
| Metabolic diseases | 8.84 × 10−76 |
| Female urogenital diseases | 3.62 × 10−74 |
| Female urogenital diseases and pregnancy complications | 2.07 × 10−73 |
| Neoplasms by histologic type | 2.79 × 10−72 |
| Neoplasms, glandular and epithelial | 1.28 × 10−71 |
| Urogenital diseases | 1.11 × 10−69 |
| Nervous system diseases | 3.04 × 10−69 |
| Pathways | Corrected p-Value |
|---|---|
| Immune system | 9.40 × 10−41 |
| Fluid shear stress and atherosclerosis | 1.39 × 10−31 |
| Innate immune system | 1.62 × 10−29 |
| Chagas disease (American trypanosomiasis) | 9.88 × 10−28 |
| Signaling by interleukins | 2.98 × 10−27 |
| Cytokine signaling in immune system | 5.26 × 10−27 |
| Apoptosis | 1.64 × 10−26 |
| AGE-RAGE signaling pathway in diabetic complications | 3.93 × 10−26 |
| Hepatitis B | 2.38 × 10−24 |
| Influenza A | 3.09 × 10−24 |
| Metabolism | 1.39 × 10−23 |
| Interleukin-4 and 13 signaling | 5.24 × 10−23 |
| Pathways in cancer | 7.23 × 10−23 |
| Pertussis | 9.62 × 10−23 |
| TNF signaling pathway | 1.08 × 10−21 |
| IL-17 signaling pathway | 4.11 × 10−21 |
| Tuberculosis | 9.85 × 10−21 |
| MAPK signaling pathway | 1.92 × 10−20 |
| Salmonella infection | 6.85 × 10−20 |
| Toxoplasmosis | 1.41 × 10−19 |
| Metal | Genotoxic Activity: Summary Report | Carcinogenic Activity: Summary Report |
|---|---|---|
| Arsenic | Positive (based on assay performed on human peripheral blood lymphocytes) | Positive (considered carcinogenic to humans by multiple health agencies) |
| Lead | Positive (assays: sperm morphology, mammalian sperm morphology test (in vivo)) | Probable/possibly/reasonably anticipated to be carcinogenic to humans by different sources |
| Negative (assays: human (in vitro) and non-human (in vivo) chromosome aberration tests) | ||
| Mercury | No information available | Mixed results (not classifiable as to human carcinogenicity by multiple health sources; classified as possible human carcinogen by one source) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ellwanger, J.H.; Ziliotto, M.; Chies, J.A.B. Toxicogenomics of Arsenic, Lead and Mercury: The Toxic Triad. Pollutants 2025, 5, 18. https://doi.org/10.3390/pollutants5030018
Ellwanger JH, Ziliotto M, Chies JAB. Toxicogenomics of Arsenic, Lead and Mercury: The Toxic Triad. Pollutants. 2025; 5(3):18. https://doi.org/10.3390/pollutants5030018
Chicago/Turabian StyleEllwanger, Joel Henrique, Marina Ziliotto, and José Artur Bogo Chies. 2025. "Toxicogenomics of Arsenic, Lead and Mercury: The Toxic Triad" Pollutants 5, no. 3: 18. https://doi.org/10.3390/pollutants5030018
APA StyleEllwanger, J. H., Ziliotto, M., & Chies, J. A. B. (2025). Toxicogenomics of Arsenic, Lead and Mercury: The Toxic Triad. Pollutants, 5(3), 18. https://doi.org/10.3390/pollutants5030018









