Abstract
Supraventricular arrhythmias (SVAs), including the often-asymptomatic supraventricular extrasystole (SVE), pose significant challenges in early detection and precise diagnosis. These challenges are of paramount importance, as recurrent SVEs may elevate the risk of developing severe SVAs, potentially resulting in cardiac weakening and subsequent heart failure. In the study conducted, an innovative approach was introduced that combined a convolutional neural network (CNN) architecture to enable the early identification and characterization of SVEs within electrocardiogram (ECG) signals. The analysis leveraged a dataset comprising 78 half-hour recordings from the highly regarded MIT-BIH Arrhythmia Database, which included annotation headers serving as labels for each recording. Signals were down-sampled by a factor of 2 and split into windows of 512 samples, with 12,288 observations for training. Following the methodology, classic signal preprocessing techniques (filtering and data normalization) were used. The proposed model was based on the UNET 1D model. A binary cross-entropy loss function, Adam optimizer, and a batch size of 128 were obtained after a hyperparameter tuning. As a training-validation methodology, a 50-fold cross-validation technique was used. The approach demonstrated a Dice coefficient of 79.01%, a precision of 80.96%, and a recall rate of 86.60% in detecting SVE events. These findings were corroborated through meticulous comparison with the annotations provided by the MIT-BIH database. The results underscore the immense potential of CNN and deep learning techniques in the early detection of supraventricular arrhythmias. This approach not only offers a valuable tool for healthcare professionals engaged in telemonitoring and early intervention strategies but also represents a significant contribution to the field of cardiac health monitoring. By facilitating efficient and precise identification of SVEs, our research sets the stage for improved patient outcomes and the prevention of severe SVAs, marking substantial advancements in this critical domain.
1. Introduction
Supraventricular arrhythmia (SVA) is a general term that encompasses several types of heart rhythm disorders that originate above the ventricles in the electrical conduction system of the heart. These arrhythmias include atrial fibrillation, atrial flutter, atrial tachycardia, and other less common forms. In fact, the prevalence of atrial fibrillation has increased three-fold in the last 50 years. Moreover, approximately 90,000 cases of supraventricular tachycardia are detected annually in the United States, and about 25% of all emergency department visits for supraventricular tachycardia result in hospitalization [1,2].
Supraventricular extrasystoles (SVEs), another type of SVA, are a common finding in persons with no pathological history specifically, with a greater proportion appearing in the elderly. They can be recognized in the electrocardiogram by the appearance of a premature narrow QRS complex, preceded or not by a P wave. This alteration originates from an electrical signal and does not produce any symptomatologic event in the person, but over time, without treatment and with greater frequency, they can trigger the weakening of the heart and subsequent heart failure [3]. Although SVEs alone are not usually dangerous, they can be a risk marker for other types of more serious supraventricular arrhythmias [4,5].
Multiple experiments have consistently demonstrated that each algorithm for detecting SVEs exhibits remarkable accuracy when applied to sampling frequencies of 120 Hz and higher [6]. In particular, the utilization of a single lead from an electrocardiogram (ECG), specifically the second lead, has proven to be exceptionally effective, obviating the need for additional leads in the monitoring process [3,7] Moreover, recent studies have demonstrated the potential of machine learning and deep learning algorithms in the detection and prediction of supraventricular arrhythmias, including those associated with supraventricular extrasystoles. For instance, a deep learning model was developed to address the identification of patients with unrecognized paroxysmal supraventricular tachycardia (PSVT) using sinus rhythm electrocardiograms [8]. The model demonstrated remarkable accuracy in detecting PSVT cases that might have otherwise been overlooked, providing a valuable tool for early diagnosis and intervention. Similarly, deep learning methodologies have been employed to diagnose ECG records under both normal conditions and supraventricular arrhythmia [9], showcasing the potential of deep learning techniques in accurately classifying different ECG patterns and distinguishing between normal and abnormal cardiac rhythms. Furthermore, innovative applications of machine learning models for the prediction of arrhythmia occurrence after acute myocardial infarction have been developed, incorporating various structural regularization techniques [10]. These studies highlight the potential of machine learning and deep learning algorithms in enhancing the detection and prediction of supraventricular arrhythmias, thereby contributing to the development of more effective and efficient telemonitoring systems for cardiac health.
Given the aforementioned considerations, the primary objective of this study was to devise a deep learning algorithm specifically designed for the identification and segmentation of supraventricular extrasystoles in ECG signals obtained from the second lead. The intended application of this algorithm is in the realm of telemonitoring for cardiac health purposes. To ensure the availability of a robust and consistent database for ECG signal extraction, the renowned and public MIT-BIH Supraventricular Arrhythmia Database was used. MIT-BIH is available online at Physionet, comprising 78 recordings of 30 min ECG data [11]. This database offers a comprehensive and diverse range of ECG readings, enabling the algorithm to be trained and tested on a wide variety of supraventricular arrhythmias, thereby enhancing its accuracy and reliability.
2. Materials and Methods
2.1. Data Acquisition
For cloud-based ECG signal extraction, the dataset of choice is the MIT-BIH Supraventricular Arrhythmia Database, which is publicly available on Physionet [11], encompassing a collection of 78 half-hour ECG records sampled at 360 Hz. This widely recognized database serves as a fundamental resource for the retrieval and analysis of ECG signals in a cloud computing environment. The dataset is composed of signals with clinical annotations (SVEs, for example), giving a wide range of future applications. By utilizing this extensive dataset, researchers and practitioners can access a diverse range of ECG recordings, enabling them to conduct in-depth investigations and develop advanced algorithms for enhanced detection and characterization of supraventricular arrhythmias. The wealth of data contained within the MIT-BIH Supraventricular Arrhythmia Database ensures the accuracy and reliability of cloud-based ECG signal extraction processes. In order to load and visualize the signals, and for the following steps in the methodology, Python programming language was used.
2.2. Preprocessing
Several signal preprocessing techniques were employed to ensure proper training. Firstly, a 50 Hz notch filter was applied to remove electrical noise. Additionally, frequencies above 150 Hz were eliminated using an FIR low-pass filter. To reduce computational costs, the entire signal was downsampled by a factor of 2, resulting in a sampling frequency of 180 Hz. Finally, signal normalization was applied, scaling the signal values from 0 to 1 to standardize the inputs for the model.
From that, the preprocessing focused on the selection of significant ECG characteristic annotations associated with EVS, including premature beats or supraventricular ectopics. These annotations were employed as labels for the subsequent stages of the project. Furthermore, the ECG signals from the MIT-BIH database were segmented into a matrix format, comprising a total of 512 samples, with each sample containing 12,288 observations.
2.3. Proposed Model
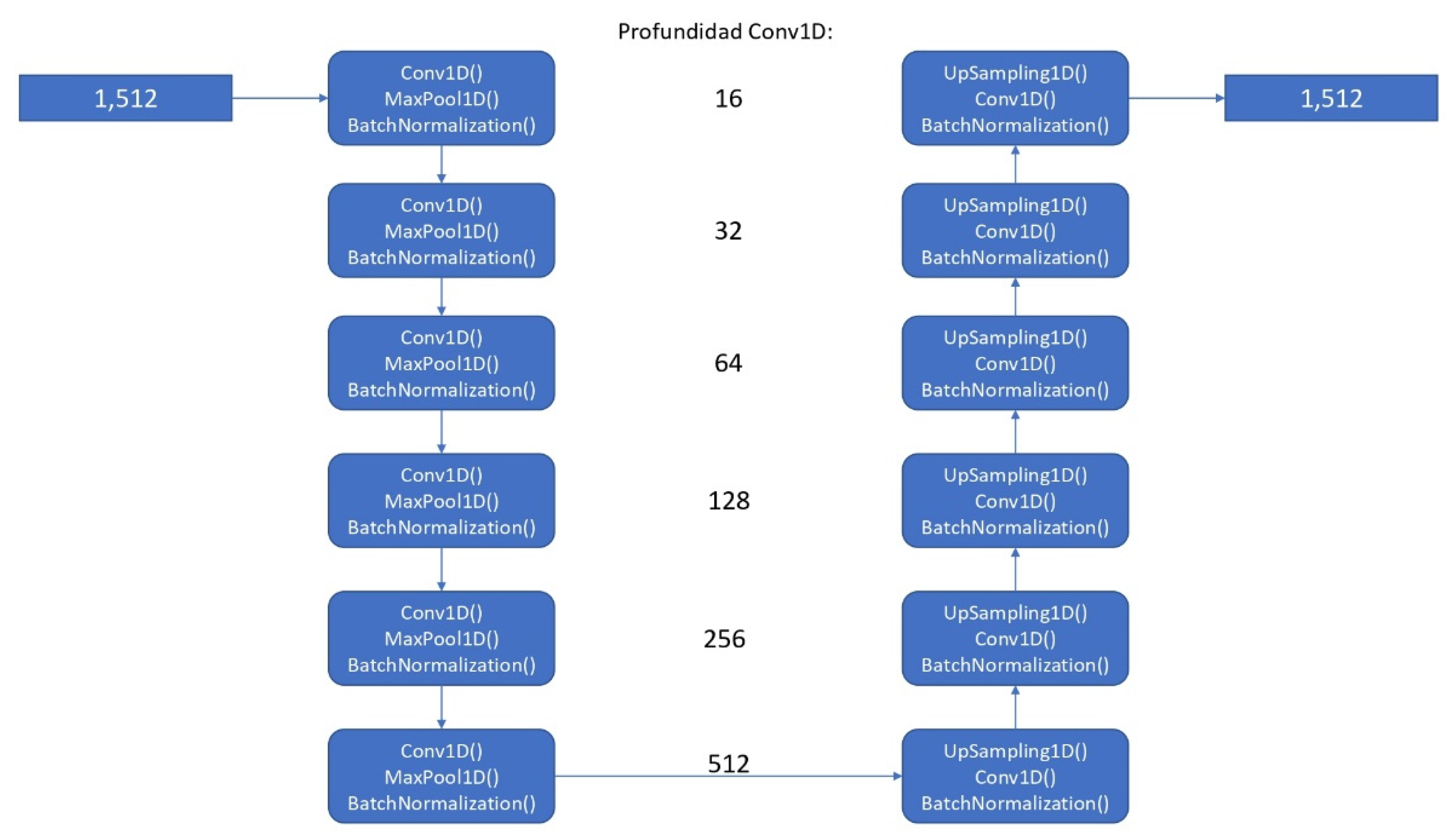
The artificial intelligence algorithm was developed using the TensorFlow open-source library, specifically utilizing a convolutional neural network model (see Figure 1) based on a UNET for 1 dimension (conv1D), taking, as a start, 64 filters, with 512 filters in last convolutional layer [12]. We considered using that approach because, for the required task (segmentation), it could be helpful to use a model that has good results in similar tasks (images) as UNET, extrapolating it to 1 dimension. As final activation function, a sigmoid function was used. We used Adam optimizer and binary cross-entropy loss function as proposed hyperparameters. As post-processing for prediction output to enhance the prediction accuracy, a threshold-based segmentation method was employed by averaging the peaks of the ECG signals.
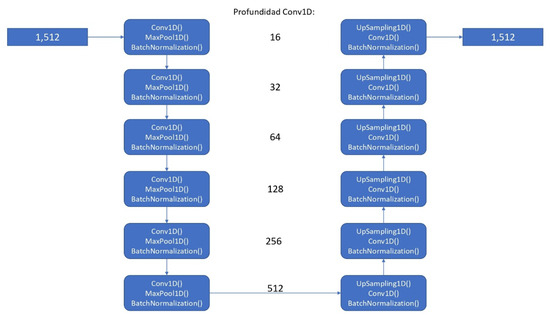
Figure 1.
Architecture of the proposed convolutional neural network model.
2.4. Evaluation Metrics
K-fold cross-validation, with k = 50, was used for training and testing of the proposed model. The average metric results will be described next. Dice coefficient, precision, and recall were used as desired metrics for evaluation.
Additionally, an interval accuracy test (called norm_accuracy, which differs from accuracy well-known metric tests) was implemented to assess the algorithm’s ability to identify true positive cases of supraventricular extrasystoles, using the norm between the difference of ground truth with predicted signal (annotations). This metric is different from the Dice coefficient because we can observe, in a more intuitive way, whether there is a good segmentation or not. The formula for this custom metric is the following:
norm accuracy = norm(prediction − ground truth)
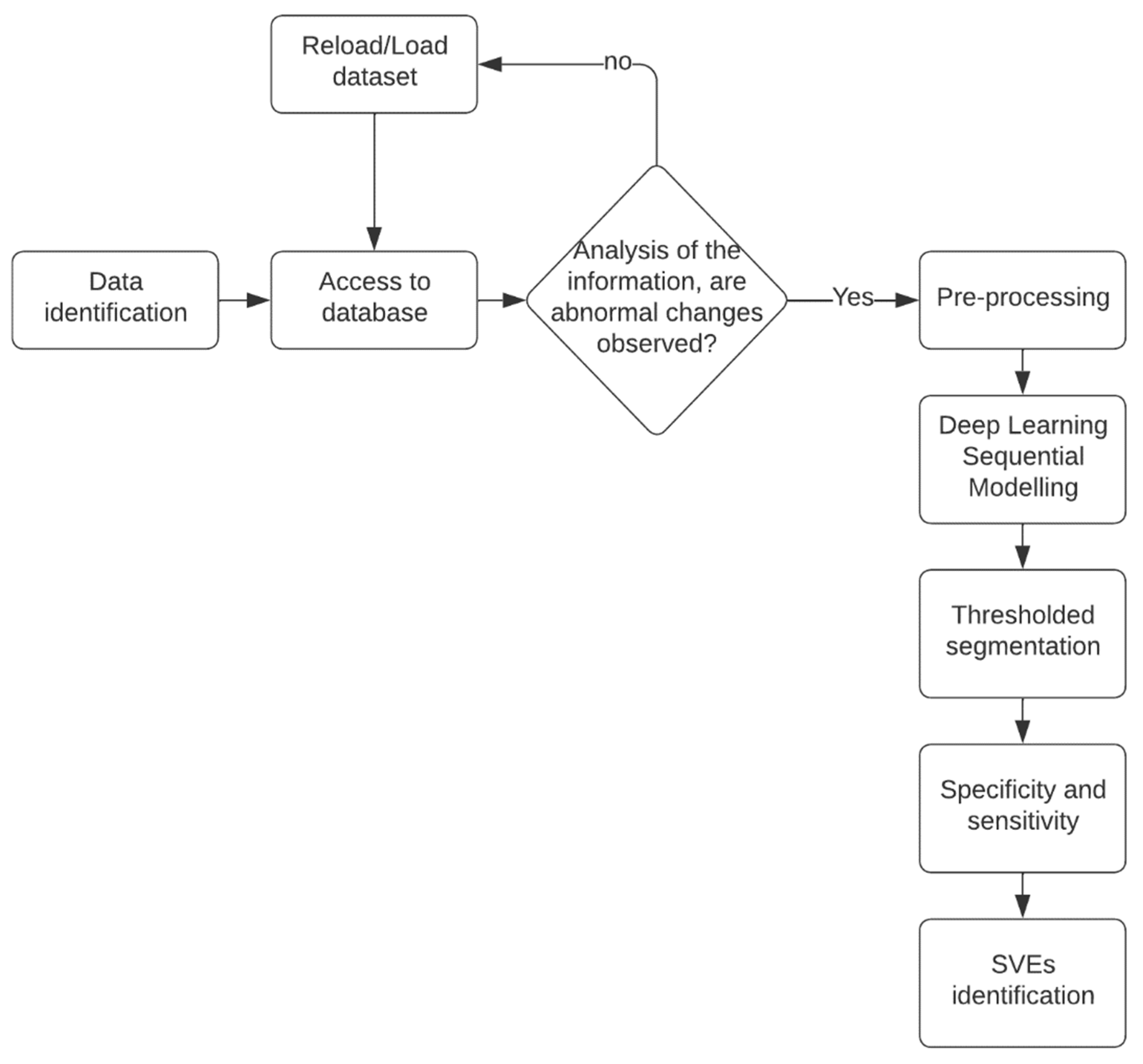
This comprehensive approach, combining data selection, preprocessing, algorithm design, and accuracy assessment, formed the foundation of the project’s methodology. Project workflow is represented in Figure 2.
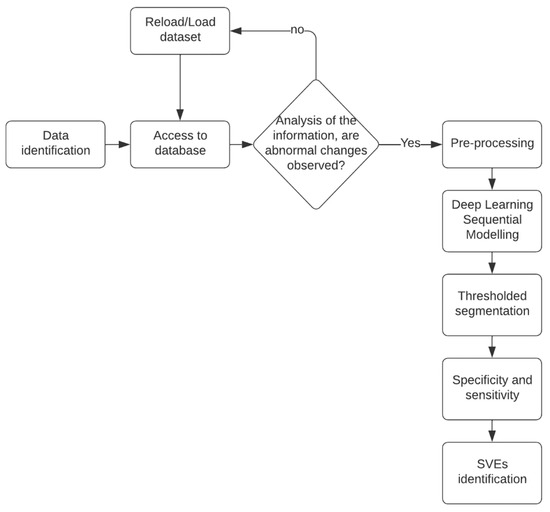
Figure 2.
Project flowchart. It starts with the choice of databases, a preprocessing of the dataset, the preparation of the neural network, its evaluation, and, finally, its validation.
3. Results
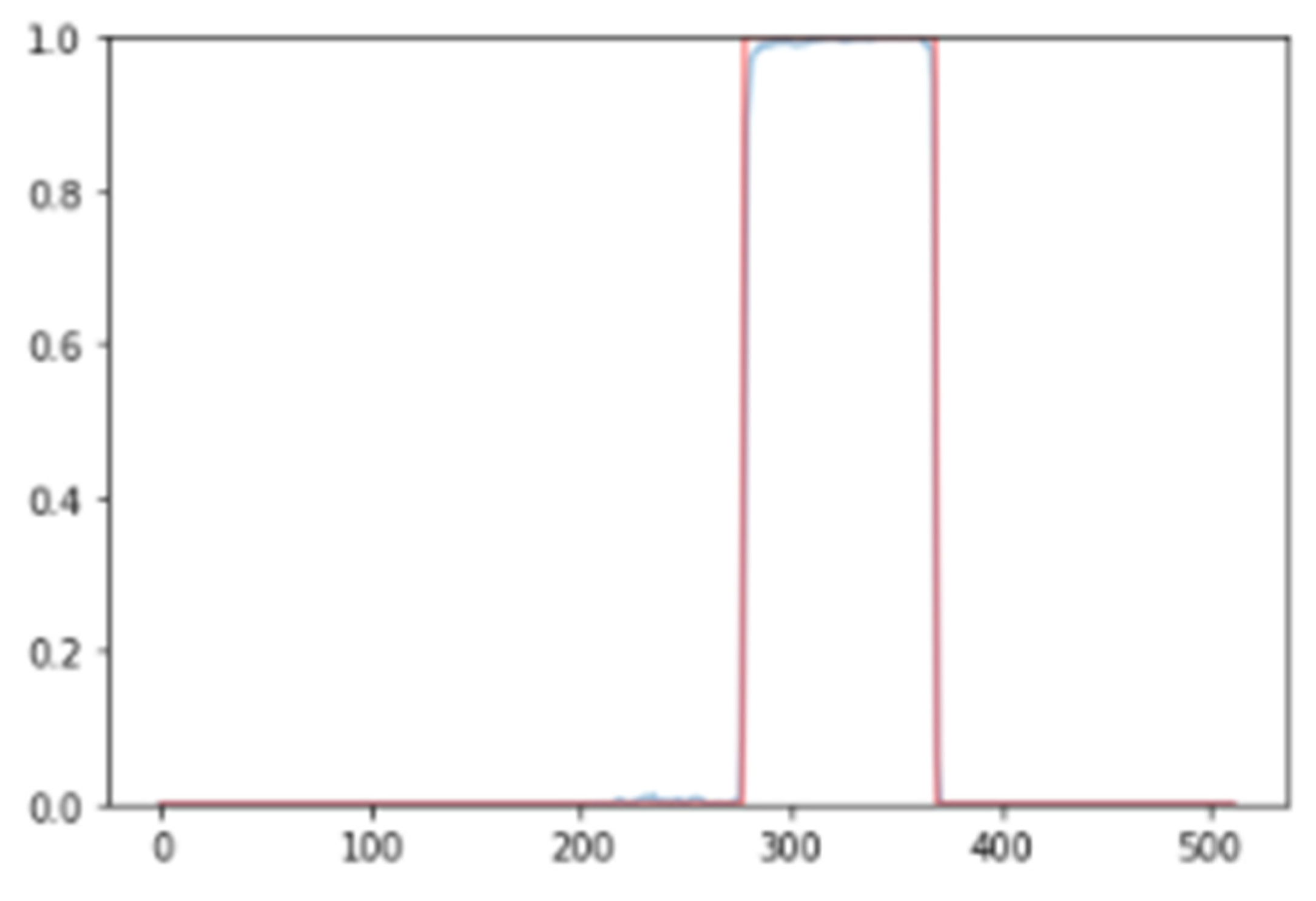
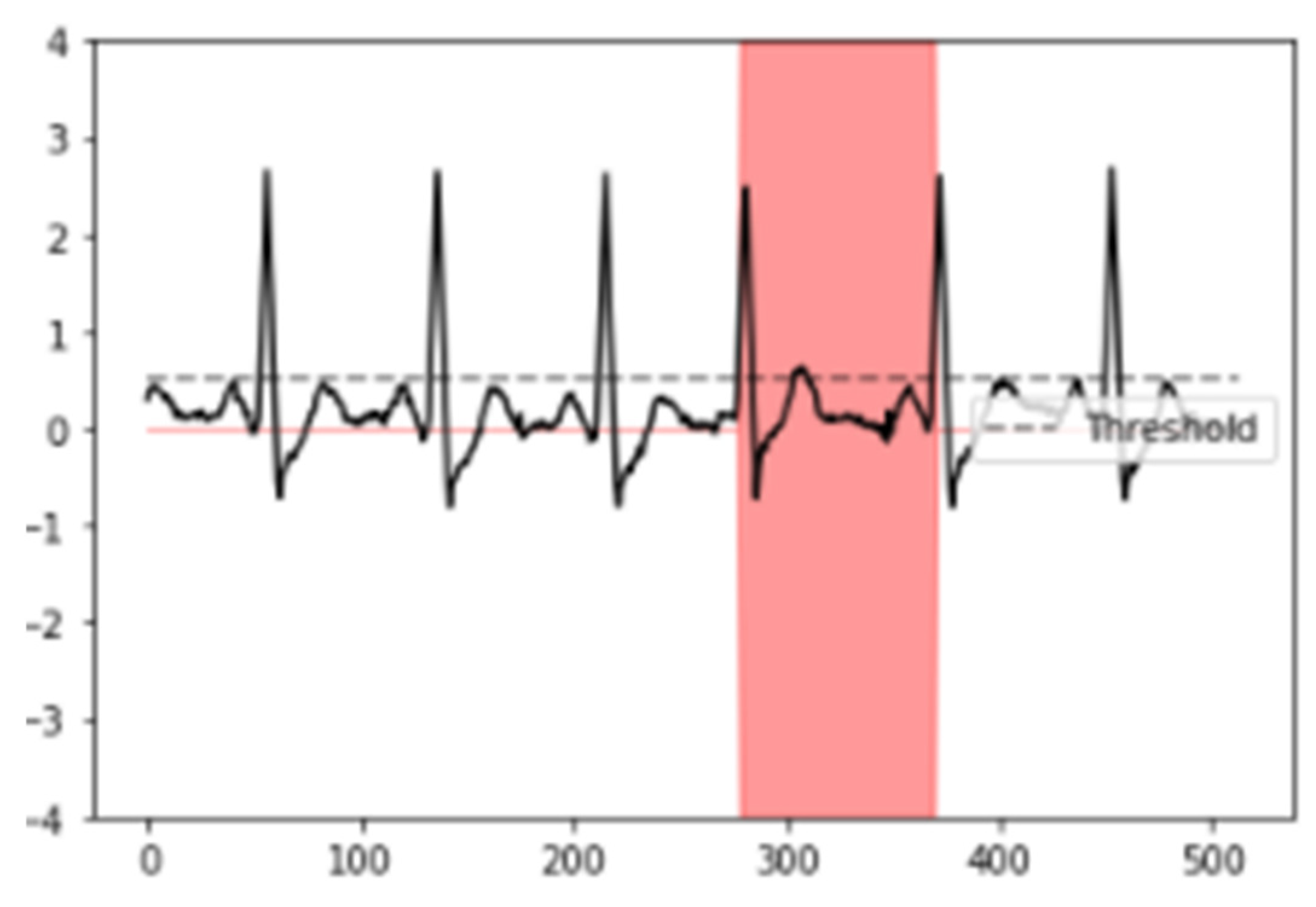
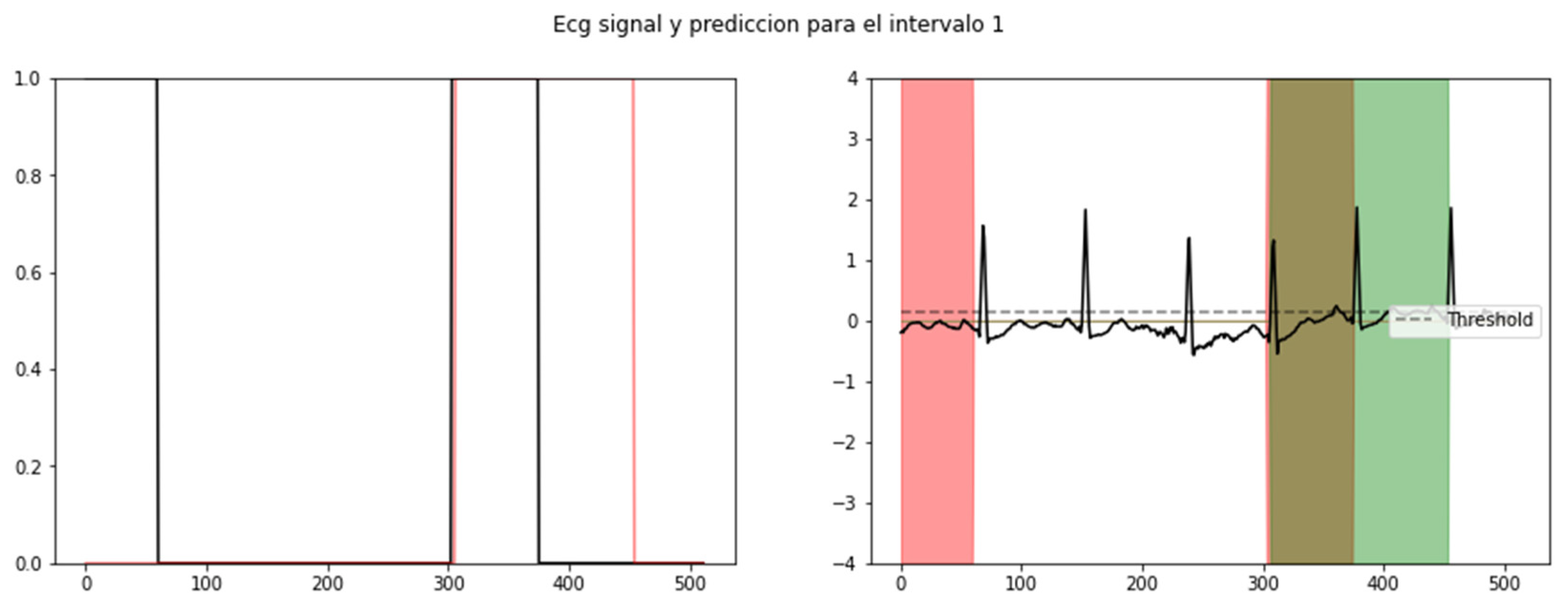
After the neural network was trained, an average Dice coefficient of 79.01% was achieved. To provide a visual representation, example results of the interval segmented and detected for SVEs and its proper ECG signal superposition are observed in Figure 3 and Figure 4, enabling the identification and visualization of sections where the R-R peak exhibited abnormal durations indicative of supraventricular extrasystoles (SVEs). The obtained results were rigorously validated against the annotations of the MIT-BIH database, confirming the neural network’s capability to accurately identify and segment supraventricular extrasystoles.
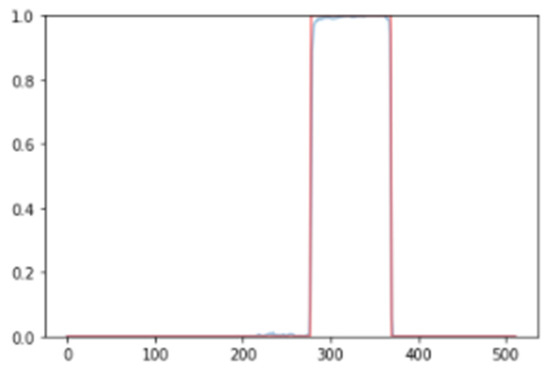
Figure 3.
Results of neural network segmentation for the detection of supraventricular extrasystole in an electrocardiogram signal. Interval detected.
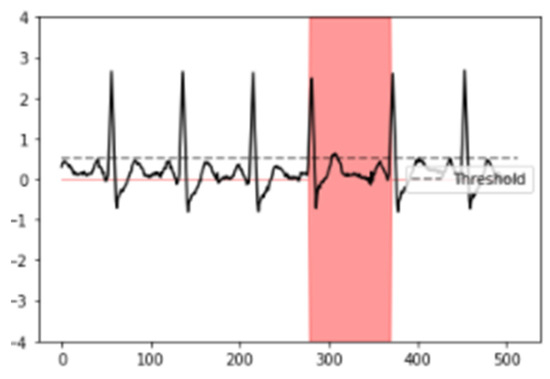
Figure 4.
Results of neural network segmentation for the detection of supraventricular extrasystole in an electrocardiogram signal. ECG signal superposed.
Following proper training of the neural network, an overall norm accuracy of 92.57% was obtained. Precision and recall were obtained with 80.96% and 86.56%, respectively, as shown in Table 1. To enhance result visualization, the Matplotlib and NumPy libraries were employed to determine a variable threshold. This threshold facilitated the complete identification of sections where the R-R peak displayed abnormal durations, indicating the presence of supraventricular extrasystoles. The validity of the results was duly verified against the annotations from the MIT-BIH database, further confirming the neural network’s proficiency in detecting and segmenting supraventricular extrasystoles.

Table 1.
K-fold cross-validation results for SVEs segmentation.
4. Discussion
The prevalence of supraventricular arrhythmias (SVAs) has been on the rise in recent years, with atrial fibrillation alone showing a three-fold increase over the past years [2]. SVAs, including supraventricular extrasystoles (SVEs), can have significant implications for cardiac health assessment, necessitating effective early detection and monitoring strategies [7]. In this study, we aimed to develop a deep learning algorithm specifically tailored for the identification and segmentation of SVEs in ECG signals obtained from the second lead, with the ultimate goal of facilitating telemonitoring applications for cardiac health.
Although good results were obtained in general for the detection of arrhythmias other than supraventricular extrasystoles, false positives can be a problem in certain cases, and efforts should be made to reduce this error. The use of functions that allow estimation of error (by normalization, for example) and precision to contrast the training dataset are therefore indispensable.
Currently, studies using artificial intelligence to support the diagnosis of supraventricular arrhythmias have not focused on supraventricular extrasystole. Despite not being the focus, the detection of SVE is useful as a marker of future more serious complications [4,5]. Therefore, our solution contrasts in methodology with the state of the art, from marker selection to the training signal and the trained model. We used only one ECG channel supported by the literature and performed a simplification of convolutional neural networks [3]. With a one-dimensional UNET architecture, we obtained results comparable with 12-channel models and with networks that are more complex [8,10].
In comparison to the state of the art, the model proposed in this study demonstrates similar overall accuracy, recall, and precision to studies detecting supraventricular tachycardia through deep learning (95.50%) and machine learning (97.00%) [8,13]. Moreover, the Dice coefficient of 79.01% indicates that there is a high degree of overlap between the predicted and actual instances of SVEs.
The results obtained from our algorithm were highly promising. The visualization of the results was facilitated by establishing a threshold, enabling the identification of sections where the R-R peak exhibited abnormal durations indicative of SVEs. As shown in Figure 5, the validation of our results against the annotations of the MIT-BIH database further confirmed the algorithm’s ability to accurately identify and segment SVEs.
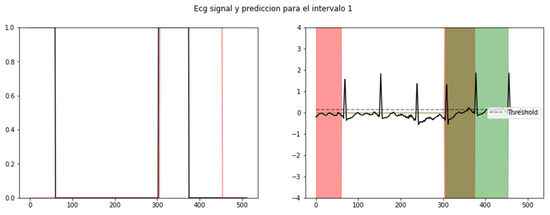
Figure 5.
Probability level of recognition of an SVE (left) and its respective segmented signal in an ECG: red (training), green (validation), and brown (coincidence) (right).
Our findings align with recent studies that have demonstrated the potential of machine learning and deep learning algorithms in the detection and prediction of supraventricular arrhythmias [8,9,10]. The utilization of deep learning methodologies, such as convolutional neural networks, has proven to be effective in enhancing the detection and characterization of these arrhythmias. By leveraging these advancements, our algorithm contributes to the development of more efficient and accurate telemonitoring systems for cardiac health.
Future research should focus on addressing this limitation and optimizing the algorithm’s performance in terms of precision. Additionally, the generalizability of our algorithm to other datasets and populations should be evaluated to assess its applicability in real-world scenarios.
In addition to its usefulness in the field of telemonitoring for cardiac health, an accurate and reliable method for the detection of supraventricular extrasystoles (SVEs) may have potential applications as a predictor of severe supraventricular arrhythmias in patients undergoing Holter monitoring. Holter monitoring, which allows continuous recording of the heart rhythm for 24 h or more, is a widely used clinical tool for assessing the presence and severity of cardiac arrhythmias. By incorporating an SVE detection algorithm into the analysis of Holter recordings, those patients at increased risk of developing more severe supraventricular arrhythmias could be identified early, allowing for more timely and personalized intervention. This predictive and early warning capability could contribute significantly to the management and treatment of supraventricular arrhythmias, improving patients’ quality of life and reducing the risk of cardiac complications.
5. Conclusions
The integration of convolutional neural networks in deep learning techniques has revolutionized the computational analysis of electrocardiogram (ECG) signals, enabling the identification and detection of previously overlooked arrhythmias, including the often-asymptomatic supraventricular extrasystole. In this research endeavor, our focus was on the implementation of a sequential convolutional neural network, along with sophisticated biological signal filters, efficient management of the MIT-BIH Arrhythmia Database, advanced processing algorithms, and an interval accuracy test to evaluate the sensitivity of SVE prediction in ECG signals. The collective findings of this study demonstrate the significant potential of our approach as a preparatory tool for future high-precision diagnostic support for SVE. By harnessing the power of deep learning and leveraging biomedical devices such as electrocardiographs for heart rate monitoring, our method can aid healthcare professionals in the early detection and accurate characterization of SVEs, facilitating timely intervention and personalized treatment strategies. This groundbreaking research opens new avenues for enhancing cardiac health monitoring and improving patient outcomes in the realm of supraventricular arrhythmias.
Author Contributions
All authors contributed equally to this research. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the MIT-BIH Supraventricular Arrhythmia Database, https://doi.org/10.13026/C2V30W.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kotadia, I.D.; Williams, S.E.; O’Neill, M. Supraventricular tachycardia: An overview of diagnosis and management. Clin. Med. 2020, 20, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Patti, L.; Ashurst, J.V. Supraventricular Tachycardia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK441972/ (accessed on 4 June 2021).
- Johnson, L.S.B.; Juhlin, T.; Juul-Möller, S.; Hedblad, B.; Nilsson, P.M.; Engström, G. A prospective study of supraventricular activity and incidence of atrial fibrillation. Heart Rhythm. 2015, 12, 1898–1904. [Google Scholar] [CrossRef] [PubMed]
- Kostis, W.J.; Ruskin, J.N. Supraventricular Arrhythmias. In MGH Cardiology Board Review; Gaggin, H., Januzzi, J., Eds.; Springer: London, UK, 2014. [Google Scholar] [CrossRef]
- Acharya, T.; Tringali, S.; Bhullar, M.; Nalbandyan, M.; Ilineni, V.K.; Carbajal, E.; Deedwania, P. Frequent atrial premature complexes and their association with risk of atrial fibrillation. Am. J. Cardiol. 2015, 116, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Ajdaraga, E.; Gusev, M. Analysis of sampling frequency and resolution in ECG signals. In Proceedings of the 2017 25th Telecommunication Forum (TELFOR), Belgrade, Serbia, 21–22 November 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–4. [Google Scholar]
- Paudel, B.; Paudel, K. The diagnostic significance of the Holter monitoring in the evaluation of palpitation. J. Clin. Diagn. Res. JCDR 2013, 7, 480. [Google Scholar] [PubMed]
- Wang, L.; Dang, S.; Chen, S.; Sun, J.-Y.; Wang, R.-X.; Pan, F. Deep-Learning-Based Detection of Paroxysmal Supraventricular Tachycardia Using Sinus-Rhythm Electrocardiograms. J. Clin. Med. 2022, 11, 4578. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, J.; Sun, L.; Cai, J.; Wang, S.; Zeng, L.; Sun, S. Application of machine learning to predict the occurrence of arrhythmia after acute myocardial infarction. BMC Med. Inform. Decis. Mak. 2021, 21, 301. [Google Scholar] [CrossRef] [PubMed]
- Bassiouni, M.; Hegazy, I.; Rizk, N.; El-Dahshan, E.; Salem, A. Deep Learning Approach Based on Transfer Learning with Different Classifiers for ECG Diagnosis. Int. J. Intell. Comput. Inf. Sci. 2022, 22, 44–62. [Google Scholar] [CrossRef]
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Volume 18, pp. 234–241. [Google Scholar]
- Howladar, P.; Sahoo, M. Supraventricular Tachycardia Detection and Classification Model of ECG signal Using Machine Learning. arXiv 2021, arXiv:2112.12953. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).