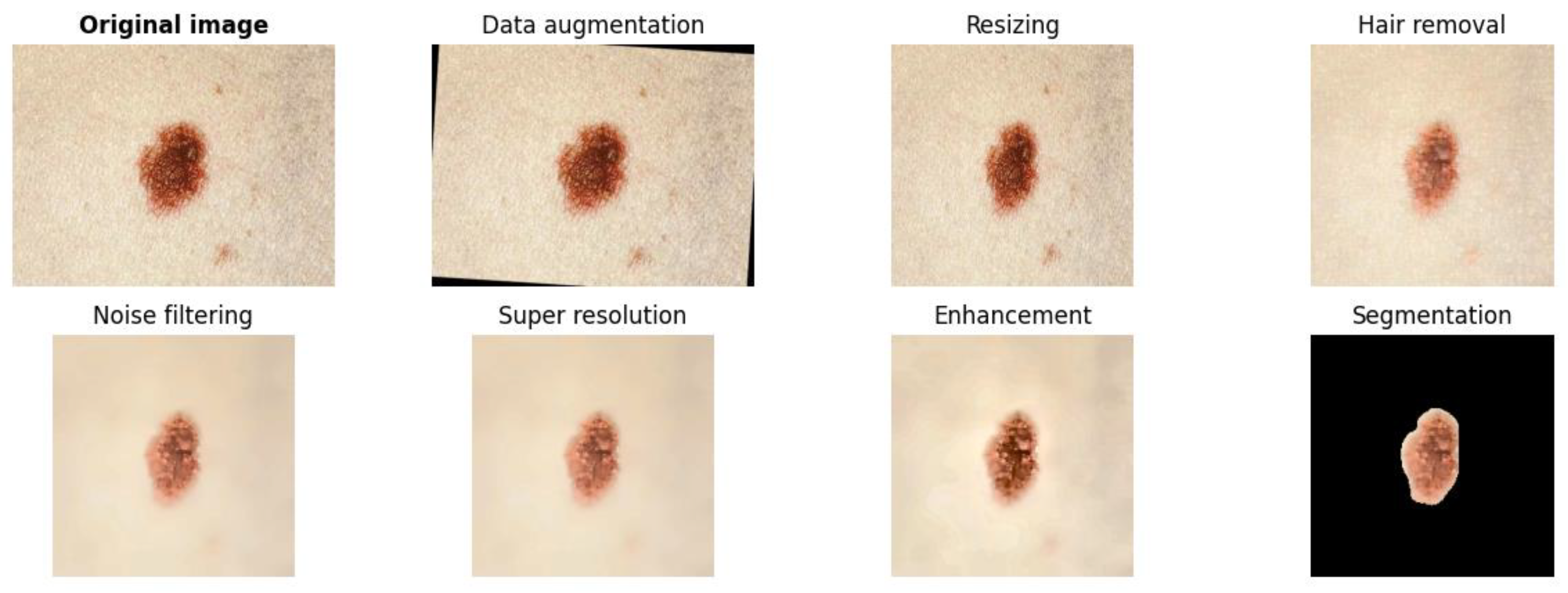
1. Introduction
The Skin Cancer Foundation [
1] indicates that dysplastic nevi, also called atypical nevi, present certain distinctive characteristics, which are grouped under the acronym ABCDE: asymmetry, irregular borders, color variation, diameter greater than 6 mm, and changes in their evolution. According to Wang et al. [
2], the existence of these nevi is associated with melanoma, which is one of the deadliest types of skin cancer in humans. This is corroborated by the study of Karaarslan et al. [
3], which has shown that having one or more dysplastic nevi generates a 2.4-fold increased risk of developing melanoma. However, Bandy et al. [
4] mentioned that the survival rate of melanoma treatment reaches 92%, if detected at an early stage. Therefore, it is important to implement systems for the detection of dysplastic nevi, which allow people to take precautions against the possible development of melanoma. However, there are several causes for the late detection of this kind of lesions, such as lack of effective skin self-examination practices, refusal of medical assistance, and misdiagnosis by a specialist.
Firstly, the lack of effective skin self-examination practices, as pointed out by Coups et al. [
5], is mainly due to insufficient guidance from health professionals on the importance and proper technique of self-examination; poor use of tools and methods that could favor a thorough evaluation of the skin, such as mirrors or the help of another person; and lack of knowledge to identify pre-malignant lesions.
Secondly, in a study concerning the reasons for delays in seeking medical attention performed on 148 patients, it was found that 38.5% of patients refused to seek medical assistance because they did not wish to have a body examination, and 9.5% of patients mentioned that they were too busy to approach a physician [
6].
Thirdly, misdiagnosis by a specialist may be due to two main reasons. On the one hand, fatigue or the mental state of the specialist may affect accuracy, resulting in an error rate of up to 20%, according to Sevli [
7]. On the other hand, dermoscopy, a noninvasive method to detect abnormalities such as dysplastic nevi [
8], is sometimes influenced by the dermatologist’s level of experience, which can negatively impact detection accuracy [
9].
Currently, there are solutions for the detection of skin lesions using clinical images collected with a smartphone, although there is still room for improvement in the results, since the accuracies of their models fail to exceed 77% [
10,
11]. This is due to the presence of many unnecessary artifacts and noise, as smartphone images are often of low quality and resolution compared to dermoscopic images [
11]. In this regard, this research proposes a model for dysplastic nevi detection using a deep learning network, designed to be implemented mainly on mobile devices. This integration with a mobile application will facilitate the use and access for people, although it can also be adapted for web platforms. The presented model considers the importance of preprocessing as a phase prior to classification, which aims to improve the quality of images that will enable better performance during medical image classification [
12]. In this context, the research uses convolutional neural networks for classification, since it has been shown to be effective in the classification of skin lesions, sometimes even surpassing the ability of expert dermatologists [
3].
In order for the proposed model to perform efficient detection, the training of the pre-trained convolutional neural network EfficientNet-B7 is limited to the use of clinical images of dysplastic nevi and common nevi, using the PAD-UFES-20 and PH2 datasets. PAD-UFES-20, which comes from the Dermatological Assistance Program (PAD) of the Federal University of Espírito Santo (UFES) in Brazil, includes a dataset of basal cell carcinoma, melanoma, squamous cell carcinoma, nevi, actinic keratosis, and seborrheic keratosis [
13]. PH2, which comes from the Dermatological Service of the Pedro Hispano Hospital in Portugal, contains a dataset of nevi, atypical nevi, and melanoma [
14].
Based on the above, the deep learning model is based on the EfficientNet-B7 architecture to detect dysplastic nevi. Such an artificial intelligence approach stands out for its high efficiency in analyzing medical images, automatically extracting increasingly complex features as the image traverses convolutional layers, addressing aspects from shape to nevus edge variation [
7,
15]. Also, it is suitable due to its low computational complexity. This architecture achieved a top accuracy of 84.3% using only 66 million parameters and 37 billion FLOPS, being significantly more efficient than other CNNs with similar accuracies. In particular, EfficientNet-B7 is 8.4 times smaller than the best GPipe model, a distributed library for training neural network models through parallelism, which underlines its efficiency in terms of parameters and computational operations, making it ideal for integration into mobile devices, as these systems have limitations in terms of resources [
16].
Mahbod et al. [
17] proposed the MSM-CNN approach, which employs multiple multiscale networks, such as EfficientNetB0, EfficientNetB1, and SeReNeXt-50, for skin lesion classification with an accuracy of 86.2%. In contrast, Baig et al. [
15] introduced Light-Dermo, an architecture based on a lightweight convolutional neural network model, using ShuffleNet-Light and preprocessing techniques to achieve an impressive 99.14% accuracy in skin lesion classification. These approaches show significant advances in skin lesion detection, highlighting the diversity of strategies and models implemented to improve accuracy.
Ou et al. [
10] developed a deep neural network with intramodality self-attenuation and cross-modality cross-attenuation to fuse image and metadata data in skin lesion classification, achieving 76.8% accuracy. In contrast, Wong et al. [
11] proposed a two-stage approach using Fast R-CNN and ResNet-50 for region-of-interest identification and binary classification, achieving a maximum accuracy of 69% by combining images and clinical data collected using a smartphone.
These approaches highlight the need to further explore other neural network architectures for better results in dysplastic nevi detection.
3. Results
In the present section, the performance of the proposed approach for dysplastic nevi detection is described in detail in order to measure the accuracy of the proposed approach and the ability to contribute to the early diagnosis of dysplastic nevi. First, the results achieved by the convolutional neural network model trained specifically for dysplastic nevi detection are presented. Subsequently, the degree of similarity between the detections made by the proposed model and the evaluation of a dermatology expert is analyzed.
3.1. Convolutional Neural Network Model Results
The results obtained using the deep learning model, together with the proposals of Ou et al. [
10] and Wong et al. [
11], are presented in
Table 1, in which superior performance by the proposed method is demonstrated in terms of accuracy, reaching a maximum accuracy of 78.33%, thus surpassing the models of Ou et al. [
10] and Wong et al. [
11], which showed lower levels of accuracy.
As can be seen, the proposal by Ou et al. [
10] focuses on the classification of six different types of skin lesions using images and meta-information. To perform this classification, they used the PAD-UFES-20 dataset, which provides a large collection of 2298 images of different types of skin lesions, including examples of melanoma, squamous cell carcinoma, seborrheic keratosis, nevus, actinic keratosis, and basal cell carcinoma, thus providing a diverse and representative basis for the development and evaluation of the proposed classification model. Another important aspect to mention is that each image had up to 21 clinical features such as age information, gender, and cancer history, among others; this approach achieved an accuracy of 76.8% and a balanced accuracy of 77.5%. In both situations, the uncertainty was 2.2%, and the metrics were calculated using the macro average.
The approach of Wong et al. [
11] focuses on the binary classification of a lesion, for the purpose of determining whether it is malignant or benign, and for the purpose of implementing an efficient model, they used three main datasets: (a) the Discovery Dataset, which includes 6819 images from 3853 patients, collected retrospectively at the Duke University Medical Center, with 57% of malignant lesions and variety in skin tones; (b) the Clinical Dataset, comprising 4130 images from 2270 patients, complemented with demographic data, lesion characteristics, and comorbidities, with 2537 images of malignant lesions and 1593 benign; (c) ISIC 2018 with 10015 images, where 1954 correspond to malignant lesions and 8061 to benign lesions. This model was able to correctly identify malignant lesions with an accuracy of 71.3% at a threshold of 0.5%, compared with accuracies of 77.9%, 69.9%, and 71.9% obtained by three board-certified dermatologists. The accuracy reported by the authors mentioned is focused on the correct classification of malignant lesions and is therefore considered to be centered in the positive class.
In this regard, the superior performance of the proposed model is largely due to three key factors: First, the implementation of various techniques to improve image quality and reduce unnecessary artifacts, such as the application of noise reduction and enhancement filters, in contrast to the approaches of Ou et al. [
10] and Wong et al. [
11], which do not incorporate such techniques. Second, the use of a convolutional neural network model, namely UNet, for image segmentation for the purpose of localizing the area of interest, which differs from the strategy of Ou et al. [
10], which does not employ any image segmentation technique, and the approach of Wong et al. [
11], which is based on the Fast-RCNN architecture. And thirdly, the choice of the EfficientNet-B7 architecture for the classification task, in contrast to the other models mentioned by authors Wong et al. [
11] and Ou et al. [
10], who used the ResNet-50 model and a fully connected layer-based classifier, respectively.
3.2. Degree of Similarity Between Expert Detection and the Proposed Model
A study was conducted that focused on evaluating and comparing two methods of detecting dysplastic nevi. One of these methods was carried out by a dermatology expert who performed a visual and clinical analysis of the nevi in sampled patients; her diagnoses were based on her medical expertise and were recorded in detail. To ensure that the dermatologist had the appropriate conditions to make accurate diagnoses, a specific protocol was followed:
Evaluation Conditions: Assessments were conducted in a suitable clinical environment, with controlled lighting, ensuring that the dermatologist had access to the necessary tools to make a detailed and accurate diagnosis.
Information Provided: During the evaluations, the dermatologist obtained information on the possible family history of skin disease through direct interviews with patients, which allowed her to contextualize each case within her clinical evaluation. Informed consent was obtained from the patients for the use of their clinical data and images in the study.
Adequate time: It was guaranteed that the dermatologist had the necessary time to perform an exhaustive analysis of each nevus without restrictions that could have affected the quality of the diagnosis.
Simultaneously, the deep learning model proposed in the study was used to detect dysplastic nevi, using digital images of the same nevi evaluated by the expert. Each nevus was photographed under controlled conditions to ensure consistency in the quality of the images and were used exclusively to compare the diagnostic effectiveness of the model with respect to the dermatologist’s direct assessment.
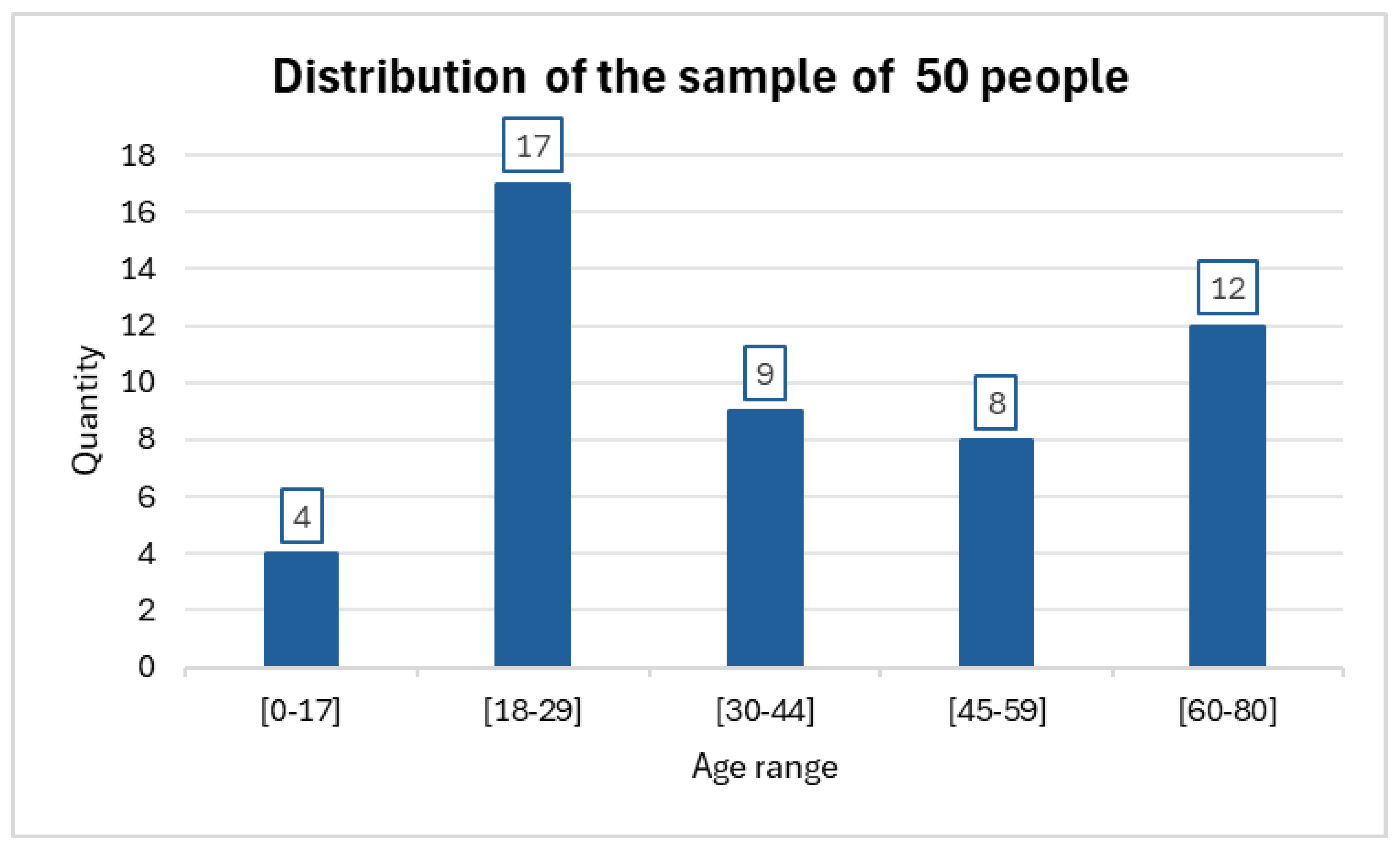
The investigation was carried out on a sample of 50 individuals who together had a total of 64 nevi. In some cases, more than one nevus was evaluated in the same patient. The distribution of the samples can be seen in
Figure 4.
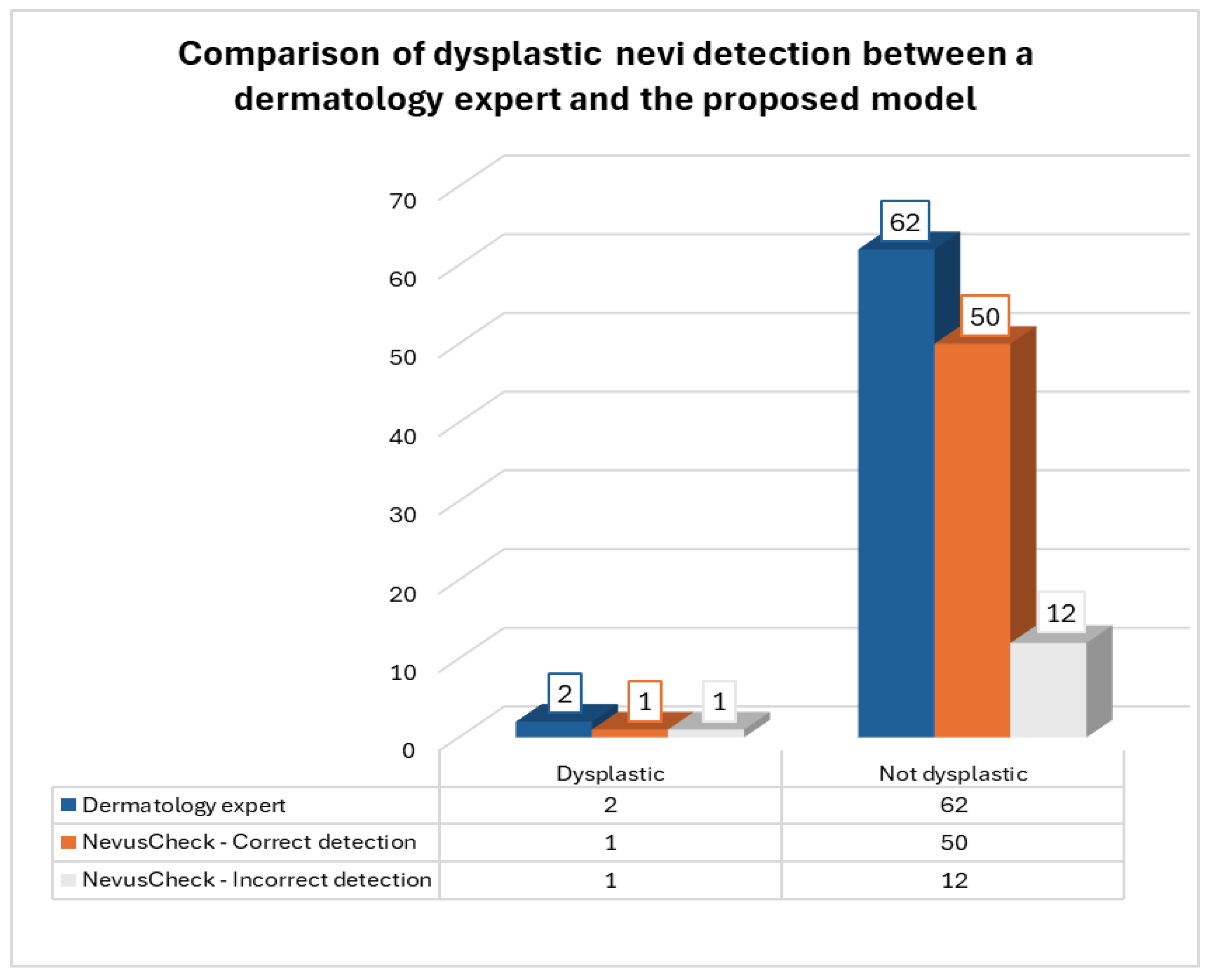
From the set of 64 nevi evaluated, the dermatologist diagnosed 2 as dysplastic and 62 as non-dysplastic. As for the NevusCheck evaluation, 13 dysplastic nevi and 51 non-dysplastic nevi were identified. In
Figure 5, it is illustrated that NevusCheck accurately identified one of two dysplastic nevi diagnosed by the expert. Moreover, the application correctly classified 50 of 62 non-dysplastic nevi as diagnosed by the expert.
Given the results, the degree of similarity between the detection by a dermatology expert and the “NevusCheck” model was 79.69%, as 51 nevi were classified in the same way out of a total of 64 nevi. This level of accuracy is remarkably close to the percentage of accuracy obtained during the validation of the deep learning model, which was 78.33%.
4. Discussion
The discussion section highlights the success of the proposed deep learning model approach focused on the detection of dysplastic nevi using convolutional neural networks.
The effectiveness of the deep learning model, with an accuracy of 78.33%, is based on known techniques such as denoising to reduce unnecessary noise and the application of the UNet model for segmentation, supporting the choice of the EfficientNet-B7 architecture for classification.
Likewise, the model performance is also based on the strategic use of PAD-UFES-20 and PH2 datasets, optimized with the data augmentation technique, resulting in a final set of 1198 images, consisting of 570 dysplastic nevi and 628 common nevi, respectively, contributing to the robust performance of the model.
In the comparison study between the evaluation of the dermatology expert and the “NevusCheck” model, a degree of similarity of 79.69% was achieved. Although the results of the deep learning model and the dermatologist differed in some cases, the proximity in the degree of similarity supports the clinical usefulness of the model as a complementary tool, considering the speed and accessibility it could offer in its implementation in mobile applications. Furthermore, it is essential to highlight that such a tool should not be considered as a replacement for consultation with a healthcare professional; rather, it acts as a complementary resource that can provide a preliminary assessment.
However, limitations in the study are recognized, especially in relation to the number of images collected with smartphones during model training. It is emphasized that the number of images of dysplastic and common nevi collected with smartphones is limited, specifically 44 dysplastic nevi images and 236 common nevi images, totaling 280 images. This highlights the need for greater focus on mobile data collection to ensure optimal application efficiency in daily use.
Future implications include extending the dataset with images acquired using mobile devices, with the purpose of increasing the accuracy achieved by the model in this study. Likewise, it is recommended to explore and evaluate different convolutional neural network structures with a view to discovering possible advances in the classification of dysplastic nevi. All of the above suggestions strengthen the ability of research to contribute to the improvement of medical care and increase skin health awareness through innovative approaches.