Abstract
Helicobacter pylori (H. pylori) is a Gram-negative, slow-growing bacterium, microaerophilic, and a unique gastric pathogen that causes chronic inflammation in the gastric mucosa with the possibility of developing gastric cancer; one-third of its proteins are still uncharacterized. In this study, computational analysis was performed on the structural, functional, and epitopic characteristics of a hypothetical protein (HP) from H. pylori named HPF63-1454. The model prediction and primary, secondary, and three-dimensional structures of the chosen HP were built. The newly created model appeared to have high quality once refinement and structure validation had been completed. The anticipated tertiary structure was assessed using the Swiss Model assessment. The best materials were chosen using structural analyses and considering data from the Z scores, Swiss Interactive Workplace, and Ramachandran plot statistics. This investigation aimed to determine the importance of the H. pylori protein HPF63-1454. Therefore, this research increased our understanding of pathophysiology and allowed us to target the protein complex specifically. H. pylori infections are now treated with readily available antibiotics and acid-suppressing medications, but this species has established itself as a hardy pest. Long-term and taxing antibiotic therapy for H. pylori infection is increasingly beginning to fail, necessitating novel and creative therapeutic modalities.
1. Introduction
Helicobacter pylori (H. pylori) is a microaerophilic, Gram-negative, spiral-shaped, slow-growing bacterium. This ubiquitous bacterium infects a substantial portion of the global populace. Australian scientists and doctors Robin Warren and Barry Marshall discovered it in 1982 [1,2]. It is an etiological agent for peptic ulcer and gastric inflammatory illnesses and a brand-new genus of bacteria that lives at the mucosa-to-lumen transition in the gut. It has the characteristics required for the colonization of the juxta-mucosal environment mucus, including microaerophilia, spiral shape, and urease positivity [3,4].
The primary cause of gastritis, gastroduodenal ulcer disease, adenocarcinoma (gastric), and mucosa-associated tissue lymphoma is H. pylori infection. H. pylori infection has drawn attention as an etiological cofactor in this cancerous illness since it is linked to an elevated risk of gastric adenocarcinoma [4]. It may disrupt a wide range of biological processes inside and outside the stomach, which could impact or determine the development of various disorders external to the stomach [5]. The most significant single risk factor for stomach malignancies is H. pylori-induced gastritis, but only a tiny percentage of infected people acquire a malignancy. Host responses, strain-specific bacterial components, and host–microbe interactions affect cancer risk. In many developing nations, the frequency among middle-aged adults is over 80%, compared to 20 to 50% in industrialized countries. The disease is acquired primarily among early childhood families and is brought on by oral consumption of the bacteria [6,7,8].
This uncharacterized protein could be helpful as a pharmacological target and marker if annotated. According to the findings of various bioinformatic tools, software, and databases, the selected protein plays a significant part in the pathogenesis of H. pylori.
2. Materials and Methods
2.1. Protein Sequence Retrieval
The selected protein’s amino acid (aa) sequence was obtained with the accession number BAW58640 (version: BAW58640.1) in the FASTA format from the NCBI [9,10].
2.2. Protein Characterization
Protein characterization was performed using the ExPASy server to measure the aa sequence composition, instability index, aliphatic index, GRAVY, and extinction coefficients. Moreover, we used the elevation of the theoretical isoelectric point (pI) for the HPF63_1454 protein [11,12].
2.3. Functional Annotation Anticipation
We utilized a conserved domain (CD) prediction in the protein of HPF63_1454. We predicted protein motif determination using the CD search functionality on the NCBI platform and the ScanProsite tool of the ExPASy server, respectively [13,14,15].
2.4. Prediction of Secondary Structure
The prediction of the secondary structure of the selected protein HPF63_1454 was anticipated using SPIPRED V.4.0 software (server-based), and the SOPMA framework was used for element anticipation [16,17].
2.5. Three-Dimensional Structure Prediction and Validation
The tertiary (3D) structure of the protein predicted by the SWISS-MODEL and the structural quality assessment studies was performed using the PROCHECK Sever of the (SAVES v.6.0) [18,19,20].
3. Results and Discussion
3.1. Protein Sequence Retrieval
Protein sequences determine the fundamental components of biological structure and function. The NCBI protein sequence database is a protein sequence database that is collected from multiple sources, including RefSeq and Gene-Bank. The protein obtained from the NCBI protein database with the accession number BAW58640 (version: BAW41740.1) was present in the locus BAW58640 containing 451 aa.
3.2. Protein Characterization
The physicochemical characteristics of the selected protein were demonstrated using the ProtParam tool. The protein HPF63_1454 contains a 451 aa long sequence with the protein that has a molecular weight (MW) of 53125.98 Da. This protein consists of 451 aa, where Leu (56) was the most abundant amino acid, followed by Lys (54), Glu (41), Asp (34), Ile (29), Ser (29), Asn (23), Thr (23), Tyr (21), Ala (20), Phe (18), Val (16), Arg (15), Pro (13), Gln (12), Gly (11), His (11), Met (10), Trp (8), and Cys (7) (Table 1).

Table 1.
Amino acid composition of the selected protein.
3.3. Functional Annotation Prediction
The NCBI CDD tool anticipated two different domains, including DUF1524 and COG1479. The DUF1524 domain is predicted to have a conserved HXXP motif that can be associated with endonuclease activities (Pfam accession: pfam07510; interval: 290–444; E-value: 2.17 × 10−18). Endonucleases cleave the phosphodiester bond that is present in nucleotide monomers. When cutting DNA, some do not care about the sequence, while others only conduct this at particular nucleotide sequences [21,22].
Moreover, The COG1479 domain contained differences between two nucleases (HNH nuclease and ParB nuclease), which was (Pfam accession: COG1479; interval: 33–188; E-value: 1.35 × 10−6). Its identical motif can be noticed in protein families and its rules as endonucleases. The actual severing of DNA strands by the nuclease domain can result in single-stranded or double-stranded breaks.
3.4. Secondary Structure Prediction
The secondary structure decisiveness of the chosen protein consensus projection from multiple alignments significantly improved the protein’s secondary structure prediction. The secondary-structural component of the protein (HPF63_1454) was a prediction made by the SOPMA software where the alpha helix (Hh), extended strand (Ee), beta-turn (Tt), random coil (Cc) was 287 (63.64%), 35 (7.76%), 7 (1.55%), and 122 (27.05%), respectively.
3.5. Three-Dimentional Structure Prediction and Validation of the Selected Protein
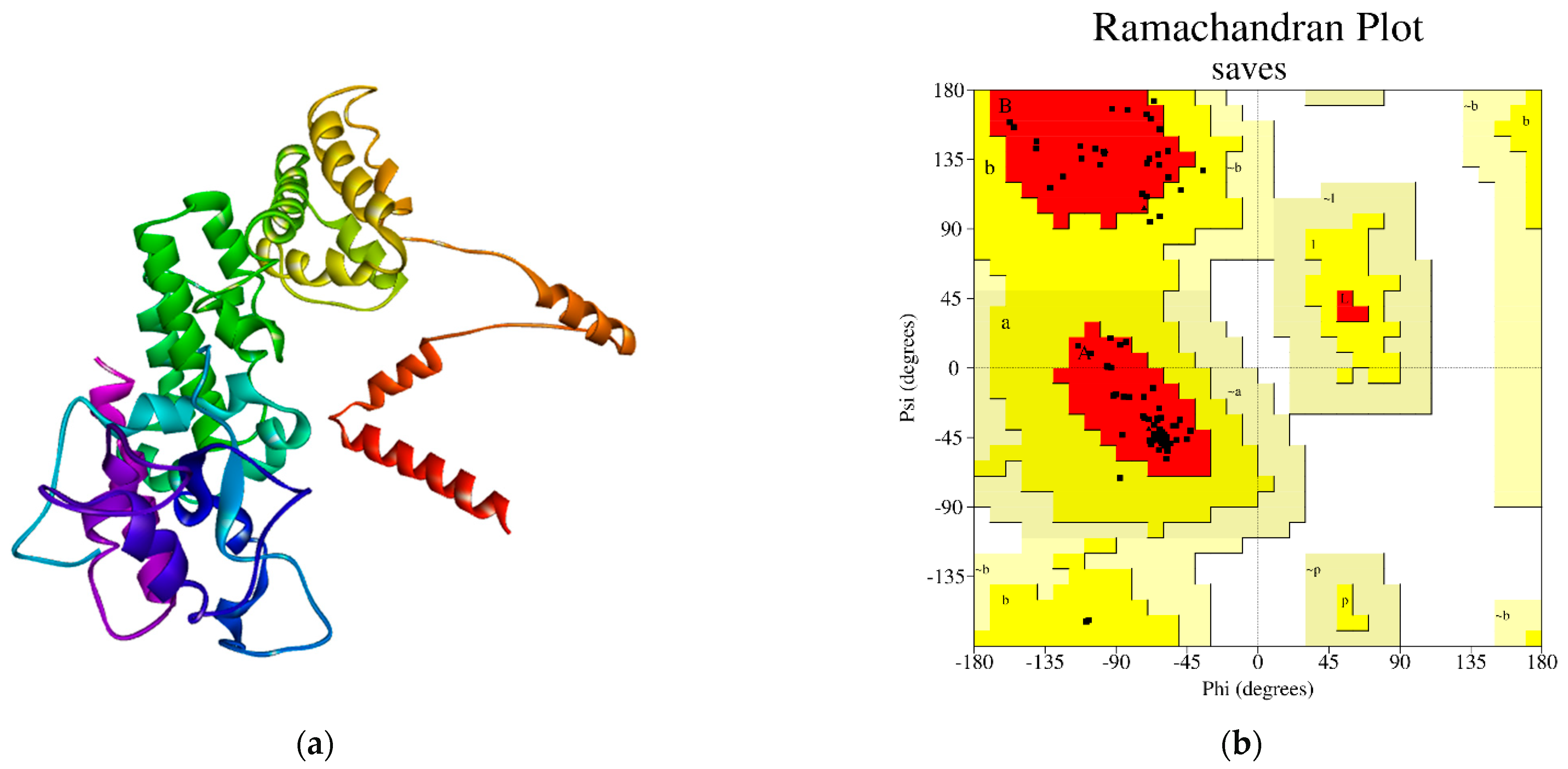
The three-dimensional architecture of the desired protein was anticipated using the SWISS-Model program [23,24]. Moreover, the PROCHECK program of the SAVES method was utilized for the structural quality assessment of the modeled protein, where the arrangement of the ψ angle and the φ angle was shown. The protein’s modeled tertiary structure was validated by the residues in the most preferred areas, which were swallowed by 92.0% of the protein (BAW58640.1) (Figure 1).
Figure 1.
(a) Three-dimensional structure modeled by the SWISS-Model tool, and (b) The PROCHECK tool, which validated the Ramachandran plot statistics of the modeled protein structure. The most favored locations are shown in red, while the other permitted zones are shown in yellow.
4. Conclusions
The bacterium Helicobacter pylori can increase the risk of developing gastric cancer. Although it is a highly deadly condition, the development of malignancy allows for a method to detect those at an increased risk daily. However, gastric infection with this bacterium is widespread, and most infected people do not proceed to catch the disease. This study used the anticipated ligand-binding active sites in Helicobacter pylori to demonstrate the structural and functional annotation of HPF63-1454. The analysis of the protein structure led to the placement of amino acid sequences in the desired location. The physicochemical characteristics and available enrichment estimation were steady in comprehending the protein operations. In most arrangements, alpha-helix, random coils, extended strands, and beta twists were found to predominate, as confirmed by the secondary assumption and assessment structures. The findings of this study may contribute to the modulation of new target identification and drug discoveries against the protein strain to reduce infections caused by the bacterium.
Author Contributions
Conceptualization, A.S.M.S.; methodology, A.S.M.S.; software, M.M.A. and A.S.M.S.; validation, A.S.M.S.; formal analysis, A.S.M.S.; investigation, A.S.M.S.; resources, A.S.M.S.; data curation, M.M.A. and A.S.M.S.; writing—original draft preparation, M.M.A.; writing—review and editing, A.S.M.S. and M.E.U.; visualization, M.M.A. and A.S.M.S.; supervision, A.S.M.S.; project administration, A.S.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request to corresponding author.
Acknowledgments
The authors would like to acknowledge the Department of Computational Biology and Bioinformatics, Advanced Bioscience Center for Collaborative Research (ABSCCR), Rajshahi 6250, Bangladesh, for supporting this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Go, M.F. Review article: Natural history and epidemiology of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2002, 16 (Suppl. S1), 3–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yamada, N.; Wu, Y.L.; Wen, M.; Matsuhisa, T.; Matsukura, N. Helicobacter pylori infection, glandular atrophy and intestinal metaplasia in superficial gastritis, gastric erosion, erosive gastritis, gastric ulcer and early gastric cancer. World J. Gastroenterol. 2005, 11, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.; Bazzoli, F.; El-Omar, E.; Graham, D.; Hunt, R.; Rokkas, T.; Vakil, N.; Kuipers, E.J. Current concepts in the management of Helicobacter pylori infection: The Maastricht III Consensus Report. Gut 2007, 56, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Parsonnet, J.; Friedman, G.D.; Vandersteen, D.P.; Chang, Y.; Vogelman, J.H.; Orentreich, N.; Sibley, R.K. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 1991, 325, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, F.; Gasbarrini, A.; Polyzos, S.A.; Kountouras, J. Extragastric Diseases and Helicobacter pylori. Helicobacter 2015, 20 (Suppl. S1), 40–46. [Google Scholar] [CrossRef]
- Polk, D.B.; Peek, R.M., Jr. Helicobacter pylori: Gastric cancer and beyond. Nature reviews. Cancer 2010, 10, 403–414. [Google Scholar] [CrossRef]
- Feldman, R.A. Epidemiologic observations and open questions about disease and infection caused by H. pylori. In H. pylori Molecular and Cellular Biology; Horizon Scientific: Wymondham, UK, 2007; pp. 29–51. [Google Scholar]
- Rowland, M.; Kumar, D.; Daly, L.; O’connor, P.; Vaughan, D.; Drumm, B. Low rates of Helicobacter pylori reinfection in children. Gastroenterology 1999, 117, 336–341. [Google Scholar] [CrossRef]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Paul, A.K.; Dey, D.; Das, R.C.; Das, M.C. In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus. Chem. Proc. 2022, 12, 21. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Islam, R.; Mahmud, S.; Imran, M.A.S.; Alam, M.S.; Masud, M.H.; Uddin, M.E. Structural and Functional Annotation of Uncharacterized Protein NCGM946K2_146 of Mycobacterium Tuberculosis: An In-Silico Approach. Proceedings 2020, 66, 13. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- de Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Uddin, M.E.; Ahmad, T.; Mahmud, S.; Imran, M.A.S.; Ahmed, S.; Alyami, S.A.; Moni, M.A. Structural and Functional Elucidation of IF-3 Protein of Chloroflexus aurantiacus Involved in Protein Biosynthesis: An In Silico Approach. BioMed Res. Int. 2021, 2021, 9050026. [Google Scholar] [CrossRef]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- Saikat, A.S.M. Computational approaches for molecular characterization and structure-based functional elucidation of a hypothetical protein from Mycobacterium tuberculosis. Genomics Inform. 2023, 21, e25. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Ripon, A. Structure Prediction, Characterization, and Functional Annotation of Uncharacterized Protein BCRIVMBC126_02492 of Bacillus cereus: An In Silico Approach. Am. J. Pure Appl. Biosci. 2020, 2, 104–111. [Google Scholar] [CrossRef]
- Nichols, N.M. Endonucleases. In Current Protocols in Molecular Biology; John Wiley & Sons: Hoboken, NJ, USA, 2011; Chapter 3, Unit 3.12. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Das, R.C.; Das, M.C. Computational Approaches for Structure-Based Molecular Characterization and Functional Annotation of the Fusion Protein of Nipah henipavirus. Chem. Proc. 2022, 12, 32. [Google Scholar]
- Saikat, A.S.M. An In Silico Approach for Potential Natural Compounds as Inhibitors of Protein CDK1/Cks2. Chem. Proc. 2022, 8, 5. [Google Scholar] [CrossRef]
- Tuncbag, N.; Gursoy, A.; Keskin, O. Prediction of protein-protein interactions: Unifying evolution and structure at protein interfaces. Phys. Biol. 2011, 8, 035006. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).