Abstract
Currently, dietary supplements contain a wide range of non-specific concentrations of testosterone and/or its synthetic analogs, substances that are not permitted and that pose a risk to public health, which puts into perspective the need to evaluate and regulate the composition of these products. The present project proposes a control tool based on the development of a biosensor using aptamers as bio-recognition elements. The aptamer is a specific sequence of oligonucleotides with can fold into unique three-dimensional structures that interact with the analyte (testosterone and analogs). Integrally, it is proposed that the aptamers are coupled with gold nanoparticles functioning as a census and signal transduction system conducing to a biosensor with high sensitivity and selectivity and rapid response. In this work, modeling and molecular docking tools were used to evaluate the folding and structural stability of the aptamers. It is essential to carry out complete in silico analysis for the bio-recognition system and to evaluate the stability of the proposed aptamers with variations in the medium, allowing one to determine the conditions and adaptations necessary for the experimental analysis, design, and operation of the biosensor. On the other hand, evaluating the affinity and identifying the types of interactions between the aptamer and analyte allows us to locate the best candidate for the proposed aptamers. The stability of a set of nine sequences with proven interaction with testosterone was evaluated under different conditions, specifically, folding temperature (8.0 °C, 20 °C, and 30 °C), [Na+] (1.0 mm M, 50 mM, and 150 mM) and [Mg2+] (1.0 mm M, 2.0 mM, 3.0 mM, and 4.0 mM), with the MFold web server, RNA Composer, and PyMOL. The affinity and molecular interaction assays were carried out between each of the aptamers and three analytes: testosterone, testosterone undecanoate, and androstenedione using Auto dock Vina, Chimera, PyMOL, and Discovery Studio. The results showing stability and conformational changes in the aptamers allow us to conclude that the aptamers (T6, T5.1, and TESS1) are compatible with the conditions used in run tests and have high affinity for testosterone, the interactions of which are mainly established through non-covalent and hydrogen bonds.
1. Introduction
In Mexico, according to the “OFFICIAL MEXICAN STANDARD NOM-251-SSA1-2009”, non-anabolic steroids are permissible substances in food supplements, but it is difficult to control the sale and distribution of these products due to easy access to national and imported food substitutes, in addition to the growing demand and incorporation of new products into the market; therefore, any supplement may possibly contain testosterone and its analogs in a wide range of concentrations and become a cause of behavioral problems (irritability and aggressiveness), personality trait drift and mood disorders (anxiety, depression), and physical effects such as baldness, cholesteric jaundice, psychotic symptoms, kidney and liver disease, coronary heart disease, and cardiovascular events. Following the above, the need to develop detection tools that are easier to use and faster to respond is addressed for application to testosterone and its analogs. The present project is a response to the lack of food supplement regulations, principally the disallowed substances and their concentrations. A large body of research has evaluated the presence and implications of steroid substances such as testosterone and their analogs for food supplements, as reported by Balasubramanian et al. [1], Zamil et al. [2], and Jędrejko et al. [3].
According to the above, this study addresses the need to develop a detection tool that is easier and quick to use for essential targets: testosterone and its analogs. We propose the development of a biosensor using aptamers as bio-recognition elements, coupled with gold nanoparticles functioning as a census and signal transduction system conducing to a biosensor with high sensitivity and selectivity, as well as a rapid response [4].
In the design of a biosensor, the bio-recognition part plays an important role; thus, the in-silico evaluation of this element is the central part of this work. Aptamers are single-chain sequences of oligonucleotides with the ability to recognize and detect small molecules with high sensitivity. The binding properties of aptamers not only vary according to the composition of their nucleotide bases but are also subject to the structural conformation that each aptamer adopts and can be modified according to the characteristics of the medium. In silico tools allow researchers to evaluate the stability of the aptamer by maintaining its conformational structure before the variation in temperature and the ion plug. Likewise, it is possible to identify the capacity and affinity of union that the aptamers present towards the molecule of interest (testosterone and analogs), its possible conformational patterns, and the type of binding links that are created between the aptamer and testosterone/analogs.
2. Methods
The aptamers considered for the development of the biosensor with proven interaction capacity were subjected to in silico evaluation for their stability and interaction with testosterone, undecanoate of testosterone, and androstenedione, which are presented in Table 1.

Table 1.
Aptamers for the design of the bio-recognition element for the biosensor.
For the structural evaluation of the nine aptamers in different conditions and each tertiary structure modeled, their affinity towards testosterone, testosterone undecanoate, and androstenedione was evaluated.
2.1. Prediction of the Secondary Structure
A library of secondary structures was generated, in CT format, based on a model of free energy minimization, applying the MFold web server [9], where the temperature conditions of 8.0 °C, 20 °C, and 30 °C were modified individually for each aptamer; the ion plug conditions, with sodium [Na+] concentrations of 1.0 mm M, 50 mM, and 150 mM and with magnesium concentrations [Mg2+] of 1.0 mM, 2.0 mM, 3.0 mM, and 4.0 mM.
2.2. Construction and Optimization of the Tertiary Structure
We then modeled the tertiary structures for each secondary structure generated using the RNA Composer web server [10], where the CT format was converted into dot-bracket format, and on the same server, the tertiary structure was obtained in .pdb format (Protein Databank File) to be displayed later with the program PyMOL.
2.3. Molecular Docking
We obtained the tertiary structures of the targets with the PubChem web server in “.sdf” format (Simulation Description Format), including those for testosterone (CID: 6013), testosterone undecanoate (CID: 65157), and androstenedione (CID: 6128). For each target, the extension was modified to “.pdb” using the Open Babel software.
After using the RNA Composer web server, the tertiary structures were obtained as RNA sequences; hence, the 3DNA-Nucleic Acid Structures web server was used to convert the structures to DNA. Later, the tertiary structure was optimized with the Avogadro software.
Molecular docking was carried out individually for each aptamer tertiary structure modeled and for each target using the Autodock Vina tool in the Chimera program [11], and the generated files were modified with Open Babel software (“.pdbqt” to “.pdb” format) to visualize the interactions and links of union between the aptamer and target using PyMOL software and Discovery Studio.
3. Results and Discussion
In the in-silico evaluation of the folding and structural stability of the aptamers, the value of ΔG was obtained. In the generation of secondary structures, the structure is considered thermodynamically stable when its value is negative, and equivalently, the numerical magnitude will denote the magnitude of stability. Consequently, the folding pattern can be visualized as conditions change, presenting more or less loops, forks, protrusions, and pseudo-knots in the spliced tertiary structure models.
Below, the ΔG value presented in Table 2 is distinguished according to the different conditions to which the modeling of each aptamer was subjected. Then, the structures tertiated using the aptamer were observed (Figure 1).

Table 2.
ΔG obtained through the modeling of the secondary structures of aptamers in varying conditions: for different temperatures in degrees Celsius (T °C), different concentrations of sodium ion in millimoles ([Na+] mM), and different concentrations of magnesium ion in millimoles ([Mg2+] mM).
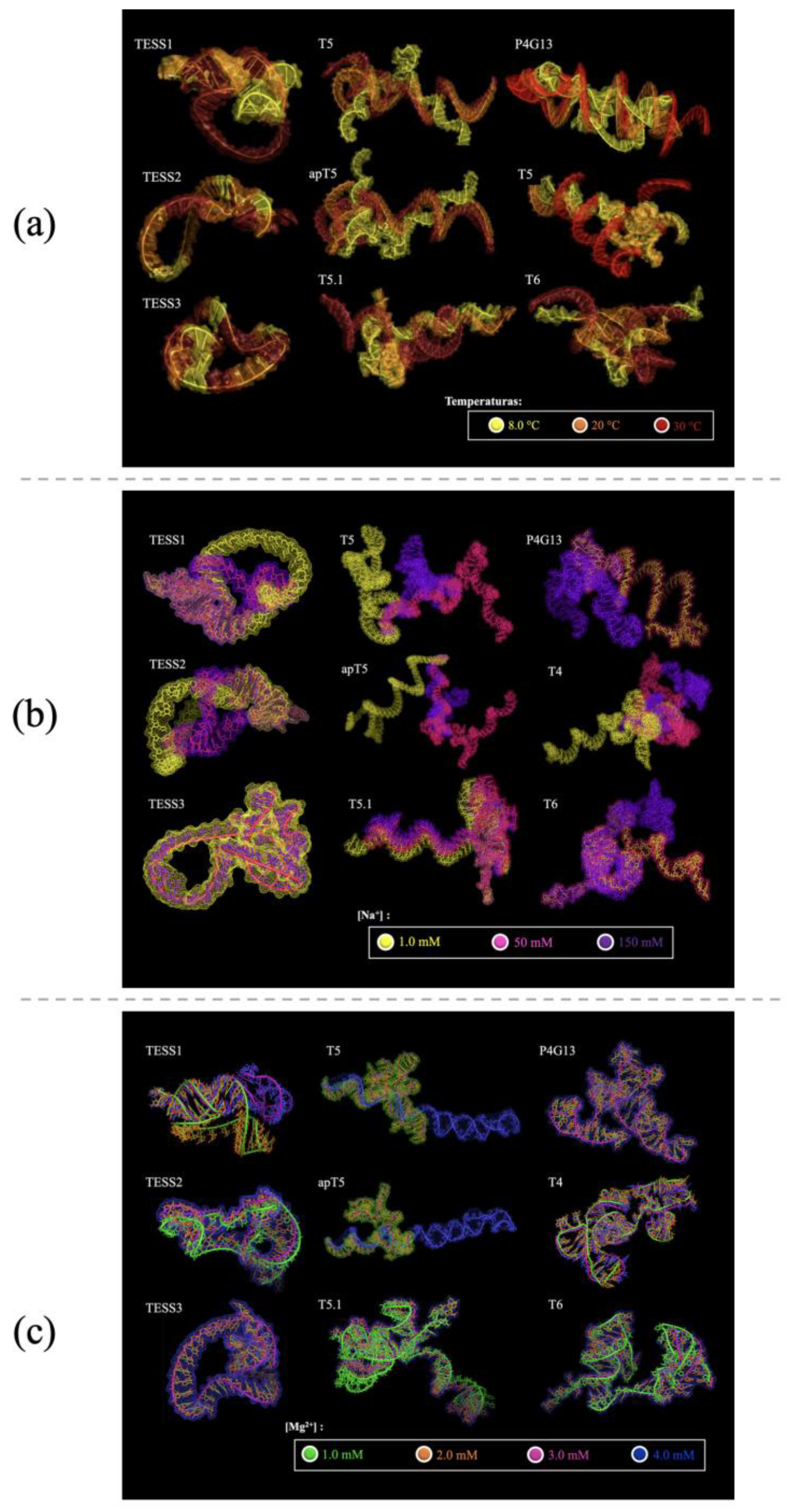
Figure 1.
Tertiary structures of the nine aptamers modeled under different conditions. (a) Different temperatures with color correspondence to 8.0 °C in yellow, 20 °C in orange, and 30 °C in red. (b) Different concentrations of [Na+] with color correspondence to 1.0 mM in yellow, 50 mM in rosy color, and 150 mM in purple. (c) Different concentrations of [Mg2+] with color correspondence to 1.0 mM in green, 2.0 mM in orange, 3.0 mM in rosy color, and 4.0 mM in blue.
According to the values presented in Table 1 above, in most cases, except for the aptamer apT5, the aptamers destabilize as the temperature increases, because at high temperatures, the bonds generated between nucleotide bases tend to separate.
At various concentrations of [Na+], the ΔG value (kcal/mol) decreases as the sodium concentration increases in response to the capacity of sodium, as a stabilizing cation for nucleic acids, to reduce the solubility that surrounds the sequence, allowing it to generate a greater number of interactions.
Evaluating the aptamers in concentrations of [Mg2+], the ΔG value (kcal/mol) decreases as the concentration of magnesium increases; that is, the higher the concentration of magnesium is, the thermodynamically more stable the structure is assumed to be. This phenomenon occurs because the magnesium ion has the characteristics of joining with the deoxyribose and forming complexes.
The relationship between Table 2 and Figure 1a, agrees with the behavior described above, in which the most compact tertiary structures present conformations with a greater number of loops and forks. It is observed in Figure 1a that some of the aptamers have almost no structural changes, such as TESS3 and TESS2, contrary to the aptamers P4G13, T5, and apT5 that were modeled at 8 °C and 20 °C and have significant changes in their structures, which decrease the possibility of adapting to the conditions in which the tests were executed. The aptamers T4, T6, T5.1, and TESS1 retain their structures between the models generated at 8 °C and 20 °C, in addition to being the aptamers with the highest negative sign values of ΔG, above ΔG = −10.00 (Table 2), suggesting that these are thermally stable aptamers.
According to Table 2 and Figure 1b, the aptamer TESS3 retains its structure at any concentration. The partially stable aptamers T6 and P4G13 have stability between concentrations 1.0 mM and 50 mM; TESS1 and TESS2 retain stability between 50 mM and 150 mM. The aptamers apT5, T4, T5.1, and T5 present noticeable changes in each concentration.
According to Table 2 and Figure 1c, the molecular models present slight structural changes; however, there are aptamers whose conformational changes are more noticeable, such as apT5 and T5. The aptamers T4, TESS1, T6, T5.1, and TESS2 present a single structural change, while P4G13 and TESS3 have no conformational changes.
Concerning the above results, we selected three aptamers with structural stability, higher values of ΔG, and less configurational changes in the different conditions. Starting with molecular docking, we focused on the selected aptamers (T5.1, T6, TESS1) and the average conditions of experimental works that we inferred would work in the future: a temperature of 20 °C, sodium ion concentration of 50 mM, and magnesium ion concentration of 2.0 mM.
According to Figure 2, the conformations of the aptamers result in a kind of cavity or pseudo-cavity, in which the target, namely, testosterone, androstenedione, or testosterone undecanoate, is already located for its union. These junctions are mostly composed of conventional hydrogen bonds, carbon–hydrogen bonds, pi–alkyl bonds, and pi–sigma bonds.
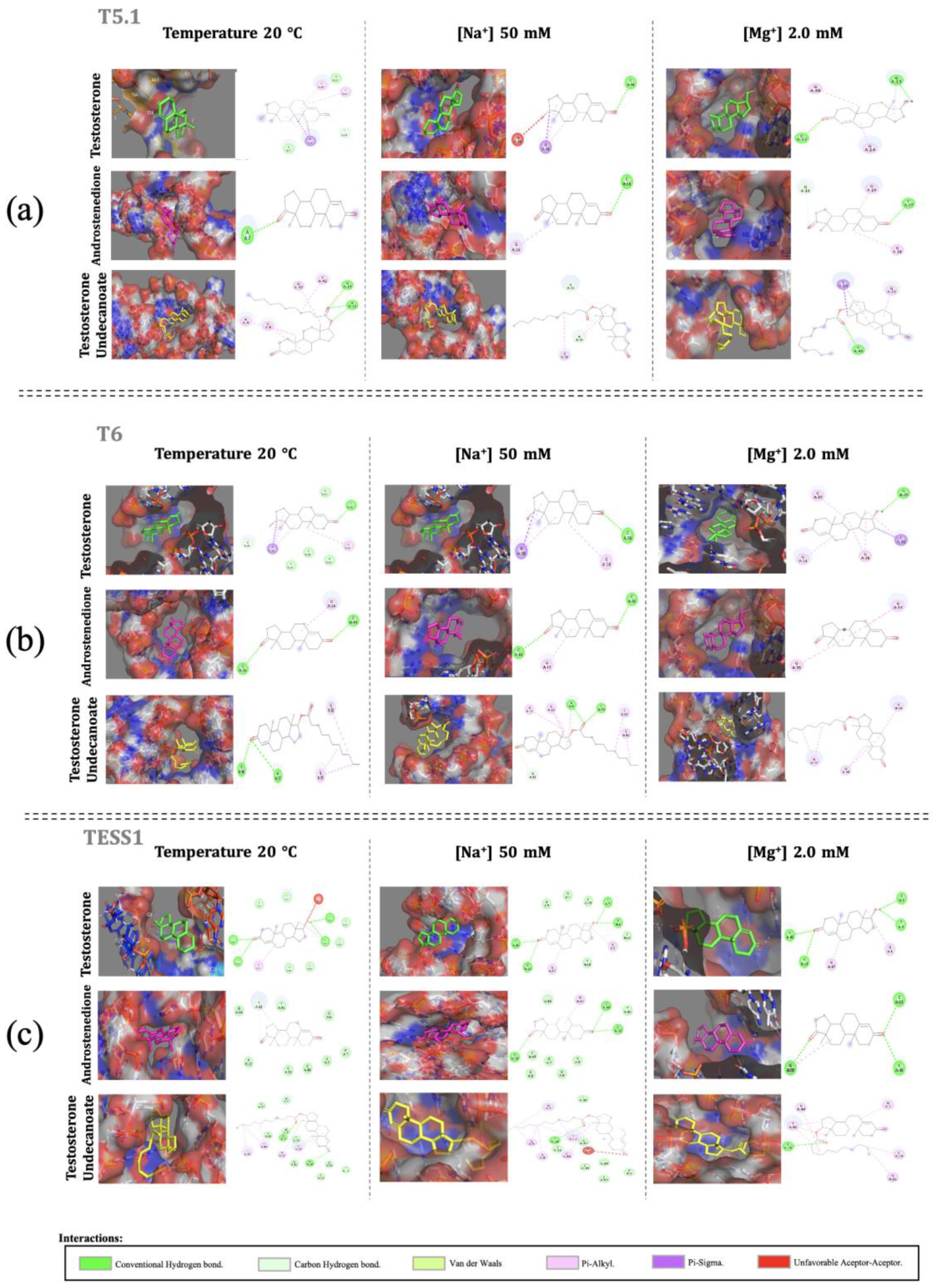
Figure 2.
Molecular docking for (a) T5.1, (b) T6, and (c) TESS1 using PyMOL images for 3D structure visualization and Discovery studio for the images of 2D structures to visualize all aptamer–target interactions.
The aptamers T5.1 and T6 have more pi–alkyl (pi–alkyl, pink) links which, despite being considered weak intermolecular forces, are interactions modulating affinity and selectivity between protein–ligand complexes, a scheme very similar to our aptamer–target and pi–sigma (purple) interactions. Conventional hydrogen bonds are another type of interaction that also occur with high incidence. These links are related to molecular recognition, being dynamic bonds, and minimize competition with water molecules. Finally, carbon–hydrogen interactions are shown to be distributed around the aptamer due to the shape of the cavity that “wraps” the target. TESS1 presents conventional hydrogen bonds, as well as van der Waals forces in greater quantity than the other types of interactions, and two situations of unfavorable acceptor–acceptor interaction, which are directly related to the conformational structure this aptamer adopts.
4. Conclusions
We discarded aptamers that did not possess thermal stability, such as the aptamer P4G13, and/or stability in the ionic plug, as for the aptamers T5, apT5, and T4, in the compatible conditions used to run the tests, performing molecular docking only with the aptamers T6, T5.1, and TESS1, where characteristic links with the aptamer–target junction were observed in greater quantity for the aptamers T6 and T5.1. This indicates that the generation of tertiary structures in different conditions directly modifies the type and number of interactions, as seen through molecular docking, which reaffirms the importance of in silico evaluations in providing a suitable biorecognition site for a biosensor.
Author Contributions
Conceptualization, A.M., A.A. and A.L.T.; methodology, A.M.; software, A.M.; validation, A.M.; formal analysis, A.M.; investigation, A.M., A.A. and A.L.T.; resources, A.M., A.A. and A.L.T.; data curation, A.M.; writing—original draft preparation, A.M.; writing—review and editing, A.M.; visualization, A.M.; supervision, A.M., A.A. and A.L.T.; project administration, A.M., A.A. and A.L.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Balasubramanian, A.; Thirumavalavan, N.; Srivatsav, A.; Yu, J.; Lipshultz, L.I.; Pastuszak, A.W. Testosterone Imposters: An Analysis of Popular Online Testosterone Boosting Supplements. J. Sex. Med. 2019, 16, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Zamil, D.H.; Perez-Sanchez, A.; Katta, R. Acne related to dietary supplements. Dermatol. Online J. 2020, 26, 2. [Google Scholar] [CrossRef]
- Jędrejko, K.; Lazur, J.; Muszyńska, B. Risk Associated with the Use of Selected Ingredients in Food Supplements. Chem. Biodivers. 2021, 18, e2000686. [Google Scholar] [CrossRef]
- Sefah, K.; Phillips, J.A.; Xiong, X.; Meng, L.; Van Simaeys, D.; Chen, H.; Tan, W. Nucleic acid aptamers for biosensors and bio-analytical applications. Analyst 2009, 134, 1765. [Google Scholar] [CrossRef]
- Pan, C.; Qiu, J.; Wang, L.; Yan, Z.; Huang, W.; Zhang, D.; Zhan, X.; Shen, G. Colorimetric Aptasensor for Testosterone Detection Based on Aggregation of Gold Nanoparticles Induced by Cationic Surfactant. Aust. J. Chem. 2020, 74, 261–267. [Google Scholar] [CrossRef]
- Skouridou, V.; Schubert, T.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’Sullivan, C.K. Aptatope mapping of the binding site of a progesterone aptamer on the steroid ring structure. Anal. Biochem. 2017, 531, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.A.; Chun, H.; Zhang, Y.; Pecic, S.; Nakatsuka, N.; Andrews, A.M.; Worgall, T.S.; Stojanovic, M.N. High-Affinity Nucleic-Acid-Based Receptors for Steroids. ACS Chem. Biol. 2017, 12, 3103–3112. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.K.; Alyoubi, A.O.; El-Shahawi, M.S.; Bashammakh, A.S.; Svobodova, M.; Betul Aktas, G.; Jauset-Rubio, M. One-Pot SELEX: Identification of Specific Aptamers against Diverse Steroid Targets in One Selection. ACS Omega 2019, 4, 20188–20196. [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Laing, C.; Schlick, T. Computational approaches to RNA structure prediction, analysis, and design. Curr. Opin. Struct. Biol. 2011, 21, 306–318. [Google Scholar] [CrossRef]
- Trott, A.J.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).