Abstract
A large amount of health and well-being data is collected daily, but little of it reaches its research potential because personal data privacy needs to be protected as an individual’s right, as reflected in the data protection regulations. Moreover, the data that do reach the public domain will typically have under-gone anonymization, a process that can result in a loss of information and, consequently, research potential. Lately, synthetic data generation, which mimics the statistics and patterns of the original, real data on which it is based, has been presented as an alternative to data anonymization. As the data collected from health and well-being activities often have a temporal nature, these data tend to be time series data. The synthetic generation of this type of data has already been analyzed in different studies. However, in the healthcare context, time series data have reduced research potential without the subjects’ metadata, which are essential to explain the temporal data. Therefore, in this work, the option to generate synthetic subjects using both time series data and subject metadata has been analyzed. Two approaches for generating synthetic subjects are proposed. Real time series data are used in the first approach, while in the second approach, time series data are synthetically generated. Furthermore, the first proposed approach is implemented and evaluated. The generation of synthetic subjects with real time series data has been demonstrated to be functional, whilst the generation of synthetic subjects with synthetic time series data requires further improvements to demonstrate its viability.
1. Introduction
Time series data are defined as a class of temporal data objects, a collection of chronological observations [1]. Time series data tend to be large in size and with high dimensionality. Time series data are characterized by their numerical and continuous nature, always considered as a whole, instead of a numerical field.
The motivation to investigate synthetic time series generation (STSG) is born from the VITALISE H2020 project [2]. One of the main objectives of this project is to provide virtual transnational access to data generated from several living labs (LLs) throughout Europe and beyond. To provide this transnational access, synthetic data generation (SDG) techniques have been incorporated into the controlled data processing workflow to generate shareable data for external researchers in compliance with the General Data Protection Regulation (GDPR) [3]. LLs are research infrastructures that enable research studies to take place in real-life environments. Those research studies generate data that are potentially interesting for the research community. However, given that these data contain human personal or sensitive information, they are stored internally in LL infrastructures and cannot be externally shared outside the original research context.
Traditionally, anonymization techniques have been used to allow sensitive data to be made publicly available whilst preserving privacy, but traditional anonymization techniques tend to suppress useful data because many of them add noise to the real data or delete attributes from them. In this scenario, SDG is presented as a game-changing anonymization technique, as it has the potential to create data without erasing potentially interesting data.
In this context, a workflow to make LL data accessible for external researchers by generating synthetic data (SD) has been proposed by Hernandez et al. [4]. As explained in the proposed workflow, SD is created with a clear purpose, enabling researchers to develop algorithms and analyses locally using it. Subsequently, the locally developed algorithms and analyses are remotely executed with the real data stored in LL infrastructures. This approach enables external researchers to conduct and validate experiments with GDPR compliance.
When evaluating SD quality, the following three dimensions are most commonly considered: privacy, utility, and resemblance. The main aim of the use case mentioned above is to remotely develop algorithms with SD to test them with real data later. Therefore, the utility dimension will be more relevant than resemblance when generating SD. Once the privacy of SD is ensured, in the context described above, more importance should be given to the utility dimension of the SD in contrast to its resemblance with real data.
2. Related Work
SDG has been gaining importance for privacy-preserving data publishing. It enables the creation of artificial data with a high statistical resemblance to real data without containing potentially private data [5]. In this context, Hernandez et al. reviewed the SDG approaches proposed as an alternative to anonymization techniques for health domain applications [6]. Furthermore, there are also studies in which STSG has been researched and used [7,8,9,10,11,12,13,14].
In 2018, Norgaard et al. [7] proposed the use of a supervised generative adversarial network (GAN), which is a variation in the originally proposed GAN approach [15]. In 2019, Yoon et al. [8] presented a time series specific GAN model, named Time-GAN, whose focus was on preserving the temporal dynamics of data. TimeGAN has later been used in the medical time series context by Dash et al. [9]. In 2020, Wang et al. [10] proposed a privacy-preserving augmentation and releasing scheme for time series data via a GAN (PART-GAN). This approach added differential privacy to the conditional temporal GAN (CT-GAN) [16], an approach that was proposed for generating videos. In addition, in 2020, an update on the Synthetic Data Vault (SDV) [11], a Python package used for generating synthetic data, added a specific model for generating time series data. This model is a probabilistic autoregressive (PAR) model, but its mathematical principles are still unpublished. In 2021, Hyun et al. [13] proposed NeuralProphet, a neural network variation in the forecasting tool Prophet [17], as a method for STSG to create synthetic diabetic foot patients. In 2022, Li et al. [14] presented the transformer-based time-series GAN (TTS-GAN) based on a transformer-encoder architecture.
Although the number of proposed STSG approaches is considerable, most of them do not consider the metadata of the subjects, as they are focused on generating highly realistic sequences of data. Furthermore, some of the approaches mentioned above transform time series data into the latent space, without analyzing the option to transform the generated synthetic time series back to the data format of the real data.
3. Proposed Approaches
This section presents two approaches for generating synthetic subjects containing temporal data. The first approach assumes that time series data do not require further transformations to ensure patient privacy. The second approach requires time series data to be synthesized. Thus, well-performing STSG techniques that consider the metadata of subjects are required. On this section, both approaches are presented on a general theoretical basis and the specific data generation models that have been used are specified in Section 4.
3.1. Synthetic Subjects with Real Time Series Data
This approach, as depicted in Figure 1, has been proposed to generate useful partially synthetic multi-subject datasets containing time series data. The approach of adding time series data to synthetically generated patients, by using synthetic tabular data generation techniques, was inspired by Schiff et al. [18], who proposed to enrich synthetic patients’ data with real time series data. In their approach, tabular data of synthetic patients are generated from a dataset, creating patients that have their illnesses labelled with diagnostic codes. Then, time series data, which are also labelled with diagnostic codes and do not have any relationship with the first dataset, are added to enrich patients’ information. This process is carried out by comparing the diagnostic codes of both datasets. Our approach improves Schiff et al.’s proposed approach, by linking (with a meaningful relationship) time series data with synthetically generated tabular subject metadata, starting from real cohort data containing such links.
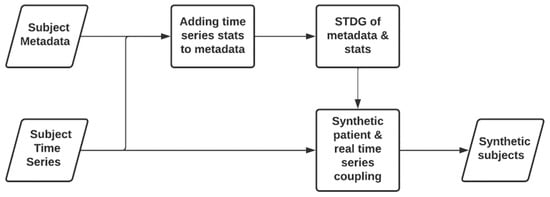
Figure 1.
Workflow for generating a synthetic patient containing real time series data.
The first and obvious step in this approach is to perform a basic exploratory data analysis. Then, data preprocessing is performed to remove missing values and inconsistencies, such as negative age values. The next step is to extract meaningful statistics from each time series and append them in a table to each subject’s metadata.
Once the multi-subject tabular data, including subject metadata and basic time series statistics, have been created, synthetic tabular data generation (STDG) techniques [6] are applied to generate a synthetic patient table with synthetic metadata and synthetic time series statistics. The last step is to couple the synthetic statistics with the statistics of real time series. This process matches each synthetic patient with the fitting time series.
3.2. Synthetic Subjects with Synthetic Time Series Data
The second approach depicted in Figure 2 could be considered the ideal approach, as its outcome is a fully synthetic dataset. This approach can be understood as an evolution of the approach introduced in Section 3.1, since it incorporates STSG techniques.
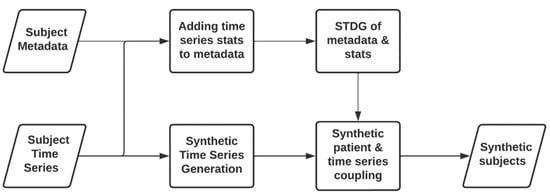
Figure 2.
Workflow for generating a synthetic patient containing synthetic time series data.
The process of generating synthetic subjects and synthetic statistics is the same as the previously explained approach. However, in this approach, time series data are synthetically generated instead of using the real time series. Once STSG techniques have been applied, the same basic statistics computed from the real time series are extracted. Considering the statistics generated with the synthetic patients and the statistics obtained from the synthetic time series data, the best fitting synthetic time series is selected for each synthetic subject, using a distance-based metric.
4. Implementation
Attempts have been made to implement the approaches presented in Section 3, but the quality of synthetic time series generated for the approach presented in Section 3.2 are not yet suitable for the target use. Thus, the approach introduced in Section 3.1, the approach that generates synthetic patient datasets and then couples them with the real time series data, has been implemented. In this section, the steps followed for the implementation of this approach are explained.
4.1. Dataset Selection
To implement the proposed approach, a dataset with a high patient volume, simple metadata, and manageable time series data that is not extremely long has been chosen. The treadmill maximal exercise tests (TMET) database [19,20] was selected from PhysioNet [21] as it fulfills the aforementioned requirements.
The TMET database is an ensemble of cardiorespiratory measurements acquired during 992 treadmill maximal graded exercise tests (GET) performed in the Exercise Physiology and Human Performance Lab of the University of Malaga. During maximal effort tests, heart rate (HR), oxygen uptake (VO2), carbon dioxide elimination (VCO2), respiration rate (RR), and pulmonary ventilation (VE) were measured on a breath-to-breath premise alongside the treadmill speed. All these measures are measured and time-stamped (Time) every 2–5 s.
The dataset is composed of two files. The first one contains all the subjects’ metadata and environmental metadata (humidity and temperature) in a tabular format. It contains data from 992 effort tests from 857 subjects, as there are subjects with several tests. These metadata are organized as shown in Table 1. The second file contains the results obtained from the treadmill experiments, i.e., the time series data. The mean length of these effort tests is 580 data rows; being each row, a measurement taken every two seconds. The time series data are organized as shown in Table 2. In this approach, each test has been considered as a subject.

Table 1.
Subject metadata variables.

Table 2.
Time series variable organization.
4.2. EDA and Data Preprocessing
Before applying the proposed approach to the selected dataset, an exploratory data analysis (EDA) has been carried out to identify missing values and inconsistencies.
Once these undesired values have been identified, a criterion to decide how to treat them has been established. If missing values are found in the time series data, a threshold of 30 data points, a 5% of the mean length of time series data, has been established to decide if the time series must be kept or rejected. Therefore, if a time series contains less than 30 missing values, imputation is made by linear interpolation. However, if more than 30 missing values are found, the time series, and the subjects they are related to, are excluded. In case missing values are found in the metadata, the exclusion of the subject, and its related time series, has been considered.
The exploratory analysis found that the environmental data were missing for 30 subjects. Those subjects were excluded from the dataset. No unexpected values were found on the metadata. The time series data analysis found that 942 HR datapoints and 4871 VO2 and VCO2 datapoints had missing values. Following the criteria described above, 12 subjects were excluded due to the amount of missing data in the time series.
Upon completing the preprocessing stage, 950 subjects remained in the dataset from the original 992 subjects.
4.3. Time Series Feature Extraction
Before selecting the appropriate statistics that need to be extracted, it is important to consider the nature of the tests from which the time series data were obtained. For the selected dataset, the data were obtained from a treadmill test. The treadmill effort test began at a speed of 5 km/h, a speed that increased 1 km/h per minute until the subject went beyond exhaustion [21]. Once the subject’s maximal effort had been reached, the treadmill speed was reduced to 5 km/h, and the recovery was recorded for 200 s.
Considering that each test has a different duration and maximal speed, the following statistics have been selected: maximal speed, test duration (last time value), and maximal, minimal, and mean values of the physiological variables (HR, VO2, VCO2, RR, and VE). These statistics have been extracted for each time series, identified with the ID_test variable, and appended to the corresponding subject in the metadata table.
4.4. Synthetic Subject Generation
The synthetic subject, and the synthetic time series statistics, have been generated by applying a STDG technique. Specifically, the Synthetic Data Vault (SDV) [11] Python package has been used. Using this approach, the generation of a cohort of synthetic subjects with their metadata and the statistics that their effort test should have were enabled. SDV contains several STDG models, from which the tabular variational auto encoder (TVAE) model with default parameters has been used to generate SD.
4.5. Time Series Coupling with Synthetic Subject
Considering that the synthetically generated statistics and the statistics extracted from the real time series do not have the same values, a strategy to couple the real effort test to the synthetic subjects is proposed. The mean value of all Euclidean distances between each synthetic time series statistics and all real time series statistics is computed. Then, the time series that brings the lowest value is selected.
Firstly, all the statistics are normalized to avoid some variables being more influential than others when calculating the mean of the distances. Subsequently, distances between each pair of statistics are calculated to compute the sum of all the distances. The ID_test of the best fitting, lowest distance sum valued, time series data are appended to each synthetic subject. Once all the synthetic subjects are assigned a time series, another dataset containing those time series is created. Finally, new identifiers linking synthetic subjects with the time series data are created. The pseudocode of this coupling can be observed in Algorithm 1.
| Algorithm 1. Coupling method. |
| def coupler(synth_row): i = ‘null’ Tid = ‘null’ for row in stats: curr = stats(row) temp = curr − synth_row dst = sum(absolute(temp)) if dst < i or i == ‘null’: i = dst Tid = stats (‘ID_test’) return Tid assigned_ID = [] for row in synth_stats: Test = coupler (synth_stats (row)) assigned_ID.append(Test) synth_info [‘ID_test’] = assigned_ID |
4.6. Validation of Subjects
To validate the metadata of the cohort of synthetic subjects, some standardized metrics and methods proposed by Hernandez et al. [22] have been used.
Firstly, the mean and standard deviation values for each variable of real data and SD have been obtained. These values are collected in Table 3. From there, it can be observed that the mean and standard deviation values of the attributes of SD are similar to those of real data.

Table 3.
Mean and standard deviation (mean ± std) values for real data and SD (where SD is generated using SDV).
A dimensionality reduction method, specifically principal component analysis (PCA), has been used to analyze whether the dimensional properties of the real cohort are preserved in the synthetic one. Figure 3 indicates that the generated cohort of synthetic subjects is quite similar in dimensionality. There are only a few points that differ from the cohort of real subjects.
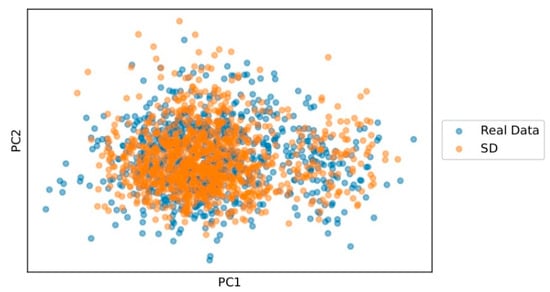
Figure 3.
PCA plot of real data (blue) and SD (orange).
Figure 4 shows the pairwise Pearson correlation (PPC) matrices for both the cohort of real subjects and the cohort of synthetic subjects. From there, it can be observed that the correlations between the attributes are very similar for both cohorts. A few correlations in the cohort of synthetic subjects are weaker than the correlations from the cohort of real subjects.
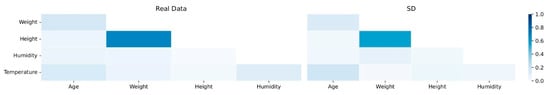
Figure 4.
Pairwise PPC matrices for real data (left) and SD (right).
A pairwise distance has been computed between each pair of real and synthetic subjects to evaluate how private the cohort of synthetic subjects is. The Hamming distance metric is used for this, which represents the proportion of attributes that are different between the two sets of records. Therefore, the higher the pairwise distance is, the better the privacy is preserved, since fewer attributes of the SD subjects are exactly equal to the attributes of the real subjects.
After computing the Hamming distance for each pair of real data and SD subjects, the distribution of those pairwise distances has been analyzed. As shown in Figure 5, most of the pairwise distance values are higher than 0.9, which indicates that for most synthetic subjects, the attributes are different from the real subject. This result indicates that privacy has been quite well preserved in the cohort of synthetic subjects.
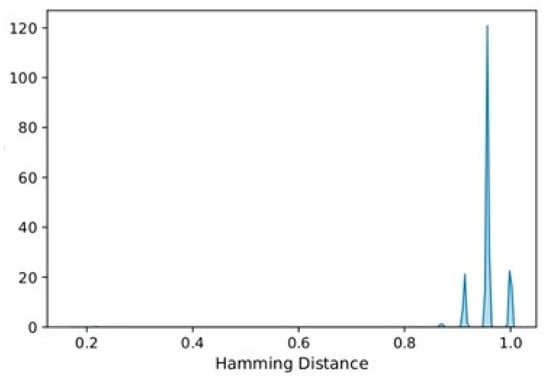
Figure 5.
Distribution of pairwise Hamming distances between real data and SD.
5. Conclusions
The main conclusion derived from this work is that the proposed approach for the generation of synthetic subjects with real time series data can be used to generate a synthetic, thus shareable dataset. Hence, it can be demonstrated that with the selected dataset, the proposed methodology can be used to generate synthetic patients and then combine them with real time series data. Furthermore, the generated cohort of synthetic subjects preserves the privacy of the cohort of real subjects, while maintaining correlations and dimensional properties.
However, the developed work has a few limitations. Firstly, more evaluation with other time series datasets should be performed to validate the generalizability of the approach used. Secondly, despite some trials implementing the second proposed approach described in Section 3.2 (using the SDV PAR for STSG), the results obtained with this model and applied to the selected dataset did not meet the minimum similarity requirements to present them. More favorable results may be obtained using this approach with other datasets. Thirdly, the privacy of the generated subject cohort has not been extensively analyzed, nor has the quality of the coupling. A more extensive evaluation of the generated synthetic cohorts should be carried out to compare different STDG or STSG techniques to select the ones that yield better results. Fourthly, this approach has been generated with only one cohort of subjects and temporal data. More research with more cohorts of subjects and time series data should be carried out to validate the generalizability and improve the approach used and the other proposed approach.
The limitations mentioned above can be taken as guidelines for future work. Firstly, other missing time series data imputation techniques, such as forecasting, will be incorporated into the data processing step. Secondly, a strategy to evaluate the coupling process will be ideated, for example, using some forecasting analysis methods. Then, the approach utilized will be evaluated with more datasets. In addition, the second approach, in which synthetic time series data are generated, will be implemented and validated, together with better performing STSG techniques and more datasets to generate multivariate time series. Concerning this approach, a method to validate the temporal nature of synthetic time series will be established, since the time series data will be fully synthetic. Furthermore, a complete strategy to evaluate the subject’s resemblance and privacy will be defined. For multivariate resemblance, the comparison of eigenvalues, the percentage of variance explained by each component and coordinates of the PCA analysis will be considered. In terms of privacy, the use of Wilcoxon signed-rank tests, the analysis of re-identification risks and computation of similarity to real data will be considered. Finally, it is intended to incorporate the proposed approaches in the VITALISE controlled data processing workflow presented by Hernandez et al. [4]. This workflow enables researchers to develop algorithms and perform analyses locally using SD and then request their execution remotely with the real data.
Author Contributions
Conceptualization, X.L., M.H., G.E. and A.B.; Data curation, X.L. and M.H.; Funding acquisition, G.E., P.B. and E.K.; Investigation, X.L., M.H. and G.E.; Methodology, G.E.; Project administration, G.E. and E.K.; Software, X.L., M.H., G.E., A.B. and C.M.; Supervision, G.E.; Validation, X.L. and M.H.; Visualization, X.L. and M.H.; Writing—original draft, X.L. and M.H.; Writing—review and editing, X.L., M.H., G.E., A.B., C.M., A.A., D.R. and E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly funded by the VITALISE (Virtual Health and well-being Living Lab Infrastructure) project, funded by the Horizon 2020 Framework Program of the European Union for Research Innovation (grant agreement 101007990).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in this study are openly available in Physionet at https://doi.org/10.13026/7ezk-j442 (accessed on 17 June 2022).
Acknowledgments
VITALISE Consortium partners and external people participating in requirements shaping open sessions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fu, T. A review on time series data mining. Eng. Appl. Artif. Intell. 2011, 24, 164–181. [Google Scholar] [CrossRef]
- VITALISE H2020. Available online: https://vitalise-project.eu/about/ (accessed on 6 March 2022).
- Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016—on the Protection of Natural Persons with Regard to the Processing of Personal Data and on the Free Movement of Such Data, and Repealing Directive 95/46/EC (General Data Protection Regulation). Available online: https://eur-lex.europa.eu/eli/reg/2016/679/oj (accessed on 22 March 2022).
- Hernandez, M.; Epelde, G.; Beristain, A.; Álvarez, R.; Molina, C.; Larrea, X.; Alberdi, A.; Timoleon, M.; Bamidis, P.; Konstantinidis, E. Incorporation of Sythetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain. Electronics 2022, 11, 812. [Google Scholar] [CrossRef]
- Emam, K.E.; Hoptroff, R. The synthetic data paradigm for using and sharing data. Cut. Exec. Update 2019, 19, 6. [Google Scholar]
- Hernandez, M.; Epelde, G.; Alberdi, A.; Cilla, R.; Rankin, D. Synthetic data generation for tabular health records: A systematic review. Neurocomputing 2022, 493, 28–45. [Google Scholar] [CrossRef]
- Norgaard, S.; Saeedi, R.; Sasani, K.; Gebremedhin, A.H. Synthetic Sensor Data Generation for Health Applications: A Supervised Deep Learning Approach. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 1164–1167. [Google Scholar]
- Yoon, J.; Jarrett, D.; van der Schaar, M. Time-series Generative Adversarial Networks. In Advances in Neural Information Processing Systems; Wallach, H., Larochelle, H., Beygelzimer, A., Alché-Buc, F.d’, Fox, E., Garnett, R., Eds.; Curran Asociates, Inc.: New York, NY, USA, 2019. [Google Scholar]
- Dash, S.; Yale, A.; Guyon, I.; Bennett, K.P. Medical Time-Series Data Generation Using Generative Adversarial Networks. In Artificial Intelligence in Medicine; Michalowski, M., Mokovitch, R., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 382–391. [Google Scholar]
- Wang, S.; Rudolph, C.; Nepal, S.; Grobler, M.; Chen, S. PART-GAN: Privacy-preserving time-series sharing. In Artificial Neural Networks and Machine Learning—ICANN 2020, Proceedings of the 29th International Conference on Artificial Neural Networks, Bratislava, Slovakia, 15–18 September 2020, Part I; Springer: Cham, Switzerland, 2020; pp. 578–593. [Google Scholar]
- Patki, N.; Wedge, R.; Veeramachaneni, K. The Synthetic Data Vault. In Proceedings of the 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Montreal, QC, Canada, 17–19 October 2016; pp. 399–410. [Google Scholar]
- Li, Z.; Ma, C.; Shi, X.; Zhang, D.; Li, W.; Wu, L. TSA-GAN: A Robust Generative Adversarial Networks for Time Series Augmentation. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; pp. 1–8. [Google Scholar]
- Hyun, J.; Lee, Y.; Son, H.M.; Lee, S.H.; Pham, V.; Park, J.U.; Chung, T.-M. Synthetic Data Generation System for AI-Based Diabetic Foot Diagnosis. SN Comput. Sci. 2021, 2, 345. [Google Scholar] [CrossRef]
- Li, X.; Metsis, V.; Wang, H.; Ngu, A.H.H. TTS-GAN: A Transformer-based Time-Series Generative Adversarial Network. arXiv 2022, arXiv:2202.02691. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N., Weinberger, K.Q., Eds.; Curran Associates, Inc.: New York, NY, USA, 2014. [Google Scholar]
- Saito, M.; Matsumoto, E.; Saito, S. Temporal Generative Adversarial Nets with Singular Value Clipping. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2849–2858. [Google Scholar]
- Taylor, S.J.; Letham, B. Forecasting at Scale. Am. Stat. 2018, 72, 37–45. [Google Scholar] [CrossRef]
- Schiff, S.; Gehrke, M.; Möller, R. Efficient Enriching of Synthesized Relational Patient Data with Time Series Data. Procedia Comput. Sci. 2018, 141, 531–538. [Google Scholar] [CrossRef]
- Mongin, D.; García Romero, J.; Alvero Cruz, J.R. Treadmill Maximal Exercise Tests from the Exercise Physiology and Human Performance Lab of the University of Malaga (version 1.0.1). PhysioNet 2021. [Google Scholar] [CrossRef]
- Mongin, D.; Chabert, C.; Courvoisier, D.S.; García-Romero, J.; Alvero-Cruz, J.R. Heart rate recovery to assess fitness: Comparison of different calculation methods in a large cross-sectional study. Res. Sports Med. 2021, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov PCh Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, M.; Epelde, G.; Alberdi, A.; Cilla, R.; Rankin, D. Standardised Metrics and Methods for Synthetic Tabular Data Evaluation. TechRxiv 2021. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).