Detection of Prostate Cancer Biomarker PCA3 with Electrochemical Apta-Sensor †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Measurements and Instrumentation
2.3. Immobilization of Aptamers and Preparation for CV and EIS Measurements
3. Results and Discussion
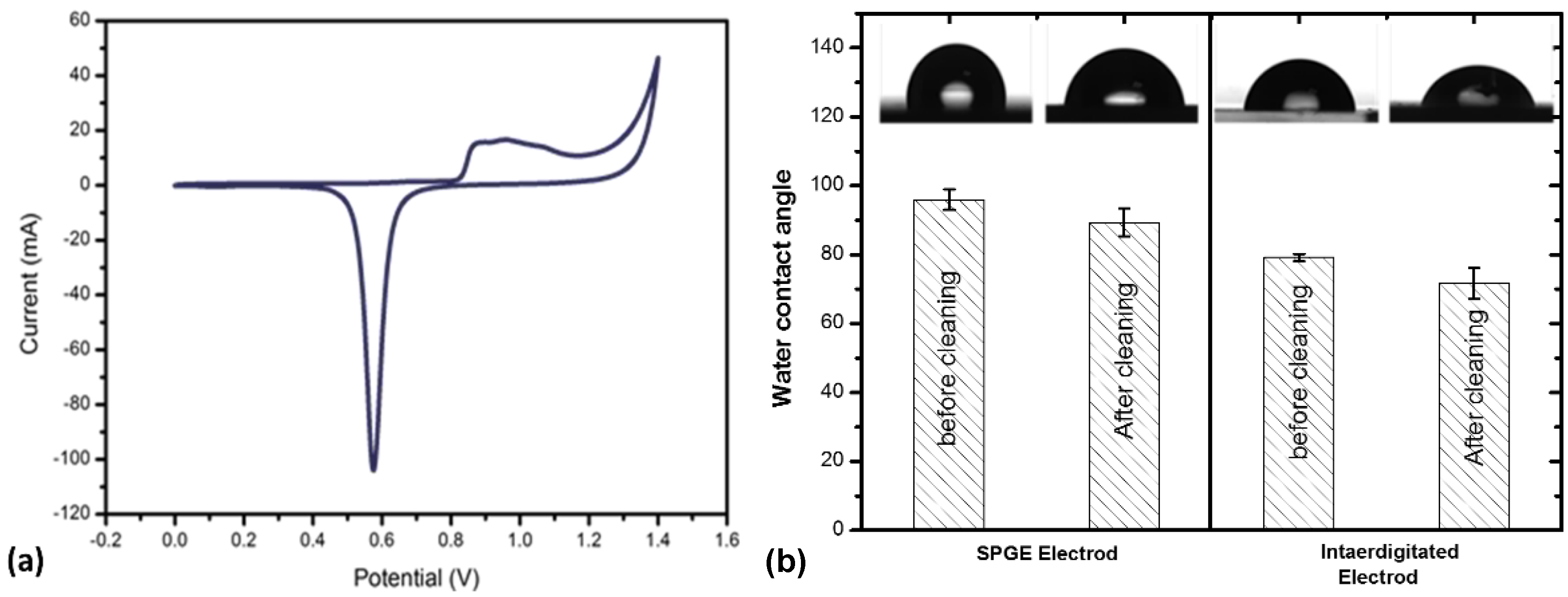
3.1. Characterization of Gold Electrodes after Cleaning
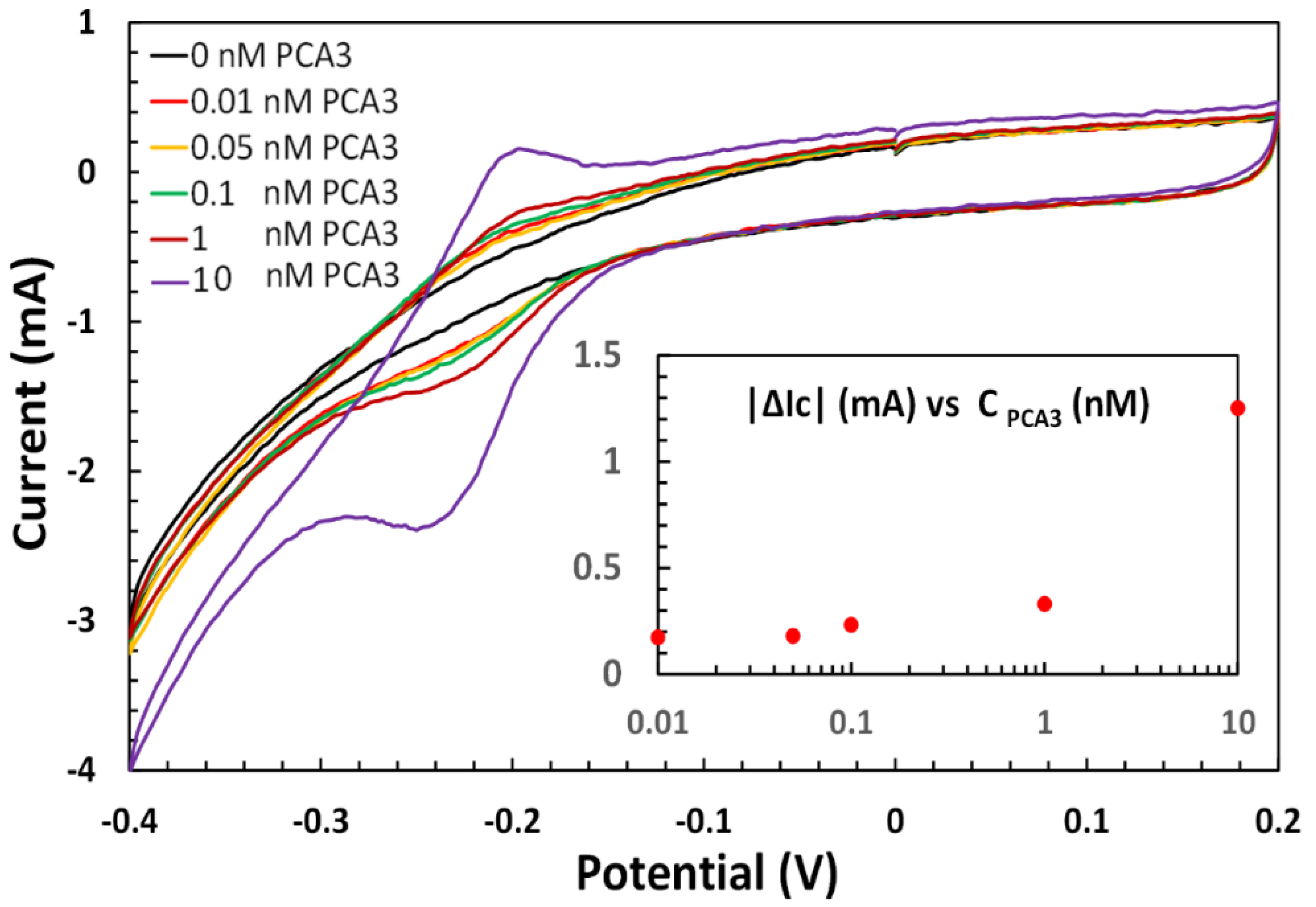
3.2. Electrochemical Apta-Sensing of PCA3
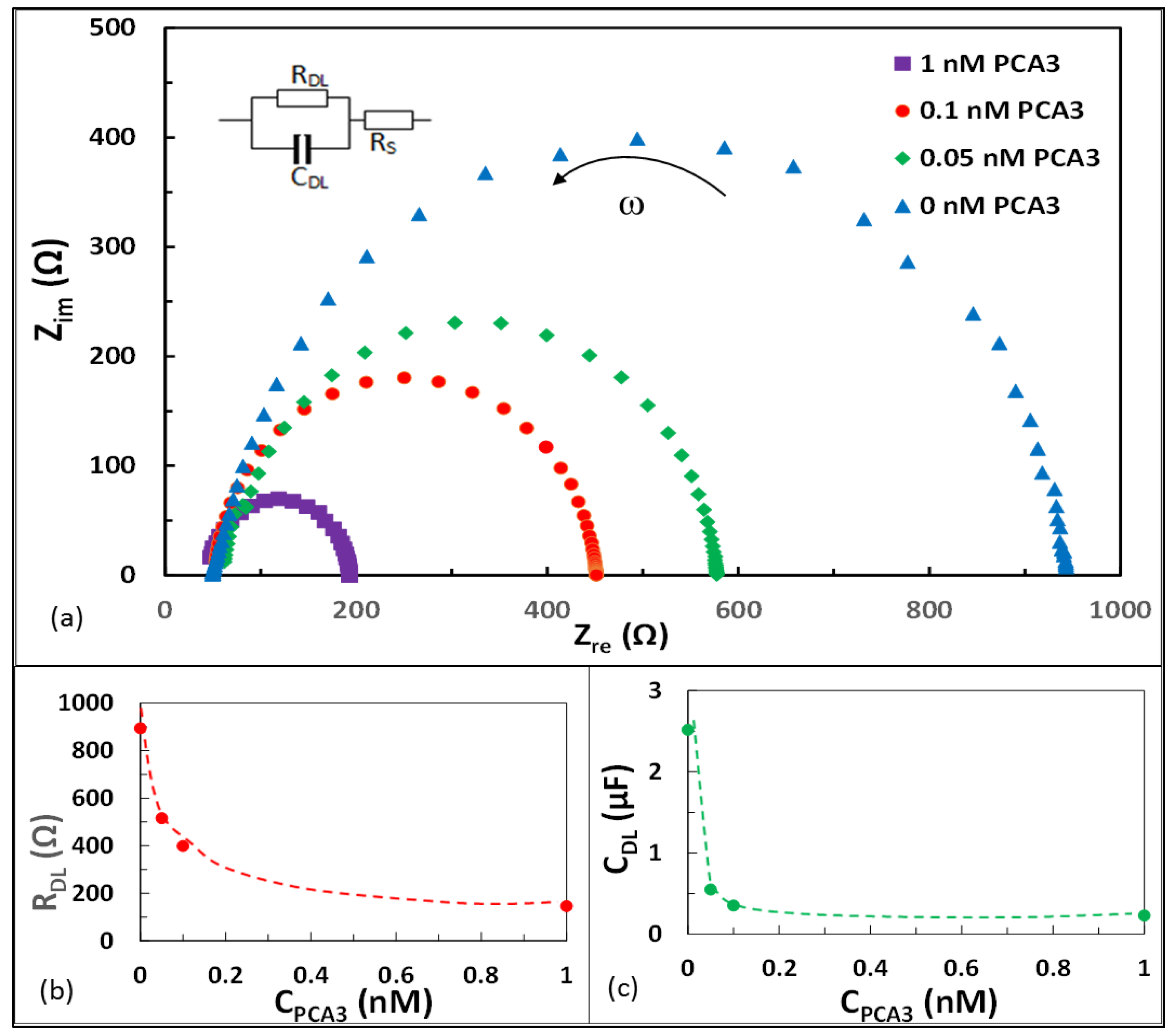
3.3. Electrochemical Impedance Spectroscopy (EIS)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Salman, J.W.; Schoots, I.G.; Carlsson, S.V.; Jenster, G.; Roobol, M.J. Prostate Specific Antigen as a Tumor Marker in Prostate Cancer: Biochemical and Clinical Aspects. In Advances in Cancer Biomarkers; Springer: Dordrecht, The Netherlands, 2015; Volume 867, pp. 93–114. ISBN 9789401772150. [Google Scholar]
- Buzzoni, C.; Auvinen, A.; Roobol, M.J.; Carlsson, S.; Moss, S.M.; Puliti, D.; de Koning, H.J.; Bangma, C.H.; Denis, L.J.; Kwiatkowski, M.; et al. Metastatic Prostate Cancer Incidence and Prostate-specific Antigen Testing: New Insights from the European Randomized Study of Screening for Prostate Cancer. Eur. Urol. 2015, 68, 885–890. [Google Scholar] [CrossRef] [Green Version]
- Daniyal, M.; Siddiqui, Z.A.; Akram, M.; Asif, H.M. MINI-REVIEW Epidemiology. Etiol. Diagn. Treat. Prostate Cancer 2014, 15, 9575–9578. [Google Scholar]
- Heijnsdijk, E.A.M.; der Kinderen, A.; Wever, E.M.; Draisma, G.; Roobol, M.J.; de Koning, H.J. Overdetection, overtreatment and costs in prostate-specific antigen screening for prostate cancer. Br. J. Cancer 2009, 101, 1833–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslan, G.; Irer, B.; Cimen, S.; Goktay, Y.; Celebi, I.; Tuna, B.; Yorukoglu, K. The Performance of Abnormal Digital Rectal Examination for the Detection of Prostate Cancer at Stratified Prostate Specific Antigen Levels. Open J. Urol. 2011, 1, 67–71. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.; Gunnell, D.; Donovan, J.; McPhail, S.; Hamdy, F.; Neal, D.; Albertsen, P.; Verne, J.; Stephens, P.; Trotter, C.; et al. Secular trends in prostate cancer mortality, incidence and treatment: England and Wales, 1975–2004. BJU Int. 2008, 101, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Mistry, K.; Cable, G. Meta-analysis of prostate-specific antigen and digital rectal examination as screening tests for prostate carcinoma. J. Am. Board Fam. Pract. 2003, 16, 95–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altuwaijri, S. Role of Prostate Specific Antigen (PSA) in Pathogenesis of Prostate Cancer. J. Cancer Ther. 2012, 3, 331–336. [Google Scholar] [CrossRef] [Green Version]
- Chistiakov, D.A.; Myasoedova, V.A.; Grechko, A.V.; Melnichenko, A.A.; Orekhov, A.N. New biomarkers for diagnosis and prognosis of localized prostate cancer. Semin. Cancer Biol. 2018, 52, 9–16. [Google Scholar] [CrossRef]
- Rönnau, C.G.H.; Verhaegh, G.W.; Luna-Velez, M.V.; Schalken, J.A. Noncoding RNAs as Novel Biomarkers in Prostate Cancer. BioMed Res. Int. 2014, 2014, 591703. [Google Scholar] [CrossRef] [PubMed]
- Schalken, J.A.; Hessels, D.; Verhaegh, G. New targets for therapy in prostate cancer: Differential display code 3 (DD3PCA3), a highly prostate cancer-specific gene. Urology 2003, 62, 34–43. [Google Scholar] [CrossRef]
- Bourdoumis, A.; Papatsoris, A.G.; Chrisofos, M.; Efstathiou, E.; Skolarikos, A.; Deliveliotis, C. The novel prostate cancer antigen 3 (PCA3) biomarker. Int. Braz. J. Urol. 2010, 36, 665–669. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.K.; Reese, A.C.; Cooperberg, M.R.; Sadetsky, N.; Shinohara, K. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis. 2012, 15, 100–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goode, R.R.; Marshall, S.J.; Duff, M.; Chevli, E.; Chevli, K.K. Use of PCA3 in detecting prostate cancer in initial and repeat prostate biopsy patients. Prostate 2013, 73, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 Molecular Urine Test: Development of a Method to Aid in the Diagnosis of Prostate Cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deras, I.L.; Aubin, S.M.J.; Blase, A.; Day, J.R.; Koo, S.; Partin, A.W.; Ellis, W.J.; Marks, L.S.; Fradet, Y.; Rittenhouse, H.; et al. PCA3: A Molecular Urine Assay for Predicting Prostate Biopsy Outcome. J. Urol. 2008, 179, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, K.; Neves, A.F.; Rocha, R.M.; Faria, P.R.; Alves, P.T.; Souza, A.G.; Fujimura, P.T.; Santos, F.A.A.; Araújo, T.G.; Ward, L.S.; et al. Prostate-specific RNA aptamer: Promising nucleic acid antibody-like cancer detection. Sci. Rep. 2015, 5, 12090. [Google Scholar] [CrossRef] [Green Version]
- Butterworth, A.; Blues, E.; Williamson, P.; Cardona, M.; Gray, L.; Corrigan, D.K. SAM Composition and Electrode Roughness Affect Performance of a DNA Biosensor for Antibiotic Resistance. Biosensors 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Jin, Q.; Jing, F.; Zhang, H.; Zhang, F.; Mao, H.; Xu, B.; Zhao, J. Integrated biochip for label-free and real-time detection of DNA amplification by contactless impedance measurements based on interdigitated electrodes. Biosens. Bioelectron. 2013, 44, 241–247. [Google Scholar] [CrossRef]
- Uludag, Y.; Narter, F.; Sağlam, E.; Köktürk, G.; Gök, M.Y.; Akgün, M.; Barut, S.; Budak, S. An integrated lab-on-a-chip-based electrochemical biosensor for rapid and sensitive detection of cancer biomarkers. Anal. Bioanal. Chem. 2016, 408, 7775–7783. [Google Scholar] [CrossRef] [PubMed]
- Takita, S.; Nabok, A.; Lishchuk, A.; Smith, D. Optimization of Apta-Sensing Platform for Detection of Prostate Cancer Marker PCA3. Int. J. Mol. Sci. 2021, 22, 12701. [Google Scholar] [CrossRef]
- Nabok, A.; Abu-Ali, H.; Takita, S.; Smith, D.P. Electrochemical detection of prostate cancer biomarker pca3 using specific rna-based aptamer labelled with ferrocene. Chemosensors 2021, 9, 59. [Google Scholar] [CrossRef]
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A Practical Beginner’s Guide to Cyclic Voltammetry. J. Chem. Educ. 2018, 95, 197–206. [Google Scholar] [CrossRef]
- Bard, A.J.; Faulkner, L.R. Electrochemical Methods: Fundamentals and Applications, 2nd ed.; John Wiley & Sons Inc.: New York, NY, USA, 2001; ISBN 978-0-471-04372-0. [Google Scholar]
- Takita, S.; Nabok, A.; Smith, D.; Lishchuk, A. Spectroscopic Ellipsometry Detection of Prostate Cancer Bio-Marker PCA3 Using Specific Non-Labeled Aptamer: Comparison with Electrochemical Detection. Chem. Proc. 2021, 5, 65. [Google Scholar] [CrossRef]
- Mussa, M.H.; Farmilo, N.; Lewis, O. The Influence of Sample Preparation Techniques on Aluminium Alloy AA2024-T3 Substrates for Sol-Gel Coating. Eng. Proc. 2021, 11, 5. [Google Scholar] [CrossRef]
- Fischer, L.M.; Tenje, M.; Heiskanen, A.R.; Masuda, N.; Castillo, J.; Bentien, A.; Émneus, J.; Jakobsen, M.H.; Boisen, A. Gold cleaning methods for electrochemical detection applications. Microelectron. Eng. 2009, 86, 1282–1285. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takita, S.; Nabok, A.; Lishchuk, A.; Mussa, M.H.; Smith, D. Detection of Prostate Cancer Biomarker PCA3 with Electrochemical Apta-Sensor. Eng. Proc. 2022, 16, 8. https://doi.org/10.3390/IECB2022-12257
Takita S, Nabok A, Lishchuk A, Mussa MH, Smith D. Detection of Prostate Cancer Biomarker PCA3 with Electrochemical Apta-Sensor. Engineering Proceedings. 2022; 16(1):8. https://doi.org/10.3390/IECB2022-12257
Chicago/Turabian StyleTakita, Sarra, Alexei Nabok, Anna Lishchuk, Magdi H. Mussa, and David Smith. 2022. "Detection of Prostate Cancer Biomarker PCA3 with Electrochemical Apta-Sensor" Engineering Proceedings 16, no. 1: 8. https://doi.org/10.3390/IECB2022-12257
APA StyleTakita, S., Nabok, A., Lishchuk, A., Mussa, M. H., & Smith, D. (2022). Detection of Prostate Cancer Biomarker PCA3 with Electrochemical Apta-Sensor. Engineering Proceedings, 16(1), 8. https://doi.org/10.3390/IECB2022-12257





