Saccharomyces cerevisiae, a yeast widely used in food and beverage production, was observed to provide a valuable source of different nutrients, the most important being the bioactive peptides which can become a major supporter in pharmaceutical therapies [1]. In this paper, the main aim was to predict in silico the health effects of bioactive peptides sourced from spent brewery yeast proteins, with a focus on their antihypertensive and antioxidant effects. To identify the bioactive peptides of interest, we employed an integrative approach which can be observed in Figure 1. We used the following tools and databases: UniProtKB, ExPASyProtParam, BIOPEP, Peptide Ranker, PepCalc and ToxinPred, and we assessed the potential of the spent brewery yeast protein and observed the presence of bioactive peptides with different effects on health [2]. The analysis of the spent brewery yeast residues of amino acids showed an increased presence of alanine, leucine, threonine, glycine and valine. Bioactive peptides were released by all three of the analysed enzymes, with the most found in the case of pepsin, followed by papain and subtilisin. All of them showed high antihypertensive activity. Using the Peptide Ranker and PepCalc tools, we managed to point out that papain and pepsin aid in the release of significant sequences such as DF, SDF, RP and RWA, which showed high scores of bioactivity and had good physico-chemical properties. In this paper, we showed that the spent brewery yeast proteins are an invaluable source of health compounds such as bioactive peptides with potential antihypertensive and antioxidant effects. The modern bioinformatics approach used in this study provided us with an efficient base for further experimental studies and offered a less costly process using databases that can simulate processes for gastrointestinal digestion.
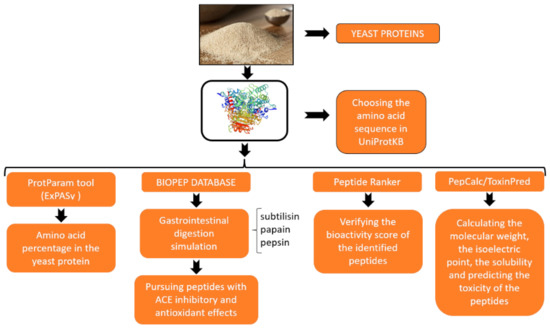
Figure 1.
The workflow used for the identification of bioactive peptides from yeast proteins.
Author Contributions
Conceptualization, F.O. and D.C.-A.; methodology, G.-M.S.; software, G.-M.S.; validation, D.C.-A.; formal analysis, D.C.-A.; investigation, G.-M.S.; resources, F.O.; data curation, D.C.-A.; writing—original draft preparation, G.-M.S.; writing—review and editing, D.C.-A.; visualization, G.-M.S.; supervision, D.C.-A.; project administration, F.O.; funding acquisition, F.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by project POC-A1-A1.2.3-G-2015-P_40_352-SECVENT, My_SMIS 105684, “Sequential processes of closing the side streams from bioeconomy and innovative (bio)products resulting from it”, subsidiary project “Plant biostimulants based on yeast extracts and hydrolysates” (1519/2019, AminAG). The SECVENT project was co-funded by European Regional Development Fund (ERDF), The Competitiveness Operational Programme (POC), Axis 1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work was funded by Subsidiary 1519/2019 of project SECVENT 81/2016 “Sequential processes of closing the side streams from bioeconomy and innovative (bio)products resulting from it”. ID:P_40_352, MySMIS:105684.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pereira, P.R.; Freitas, C.S.; Paschoalin, V.M. Saccharomyces cerevisiae biomass as a source of next-generation food preservatives: Evaluating potential proteins as a source of antimicrobial peptides. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4450–4479. [Google Scholar] [CrossRef]
- Langyan, S.; Khan, F.N.; Yadava, P.; Alhazmi, A.; Mahmoud, S.F.; Saleh, D.I.; Zuan, A.; Kumar, A. In silico proteolysis and analysis of bioactive peptides from sequences of fatty acid desaturase 3 (FAD3) of flaxseed protein. Saudi J. Biol. 2021, 28, 5480–5489. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).