1. Introduction
Polymeric membrane electrodes offer numerous advantages, and their properties can be tuned in a wide range by the modification of membrane composition. However, the process of the selection of an appropriate ligand to construct the sensor for a particular task is time-consuming and requires ligand synthesis, sensor preparation, and characterization. It would be very helpful for researchers to make a model that allows the prediction of sensor properties of an electrode based on the structure of the employed ionophore.
Using the computational chemistry, different characteristics of chemical compounds can be predicted. Quantitative relations between physical or chemical properties of organic compounds and their chemical structures can be set with the QSPR (quantitative structure property relationship). This methodology is widely applied nowadays, e.g., in pharmaceutical investigations; specifically, a search for new drugs [
1]. There are QSPR models for various materials, such as nanomaterials [
2], catalysts [
3], and ionic liquids [
4]. Recently, an application of QSPR for predicting the sensor properties of membrane electrodes was suggested [
5]. It was possible to relate the structure of the organic ligand with the selectivity constant of the corresponding membrane sensor for Ca
2+/Mg
2+.
This study aims to expand this approach to anion-selective sensors, where ligand selection is much more challenging than in the case of metal cations, due to the wide variability of the geometries of inorganic ligands, and their hydrolysis in the case of weak acids.
Among anions, there are ones with important biological and industrial roles. An example of such anions is carbonate. Therefore, it is a prospective object for predicting selectivity to carbonate against chloride anions by means of QSPR.
2. Materials and Methods
2.1. Dataset
The dataset of 40 samples was composed with literature sources and experimental data. A summary table with compositions of all samples and literature sources can be found in the
supplementary materials. Whereas the number of anionic (especially carbonate) ionophores is small due to the reasons that we have already discussed earlier, there are a few articles about carbonate ionophores. A great part of the data was extracted from IUPAC review “POTENTIOMETRIC SELECTIVITY COEFFICIENTS OF ION-SELECTIVE ELECTRODES’’, with a summary table of anions existing in 2002 [
6]. Considering the significant shortage of carbonate ionophores, we had to add in the table ionophores with Cl
−/HCO
3− selectivity that were converted to HCO
3−/Cl
− selectivity according to the Nikolsky–Eisenman equation:
We also made sure that all these data were obtained in the narrow range of pH (7.0–8.6) for understanding which particular ionic form prevailed in an examined solution.
All the membranes described in the literature are based on polyvinylchloride (PVC) as a polymer, and one of three plasticizers: nitrophenyl octyl ether (NPOE), dioctyl adipate (DOA) and dioctyl sebacate (DOS), which is also known as bis(2-ethylhexyl) sebacate (BEHS). Ion-exchanger is tridodecylmethylammonium (TDMA) with Cl− or NO3-counter-ion.
Due to the small number of carbonate ionophores examined, we had to compile the resulting database with membranes that differ not just in ionophores, but plasticizers as well. We took different plasticizers into account and added their dielectric constant (also known as relative permittivity) as a descriptor for adjusting our model and making it more comprehensive.
The selectivity coefficients of these ionophores varied from −5.8 to 6.2 on the logarithmic scale. The average selectivity logK (HCO3−/Cl−) was −1.425, and the median value was −2.6.
2.2. Descriptors
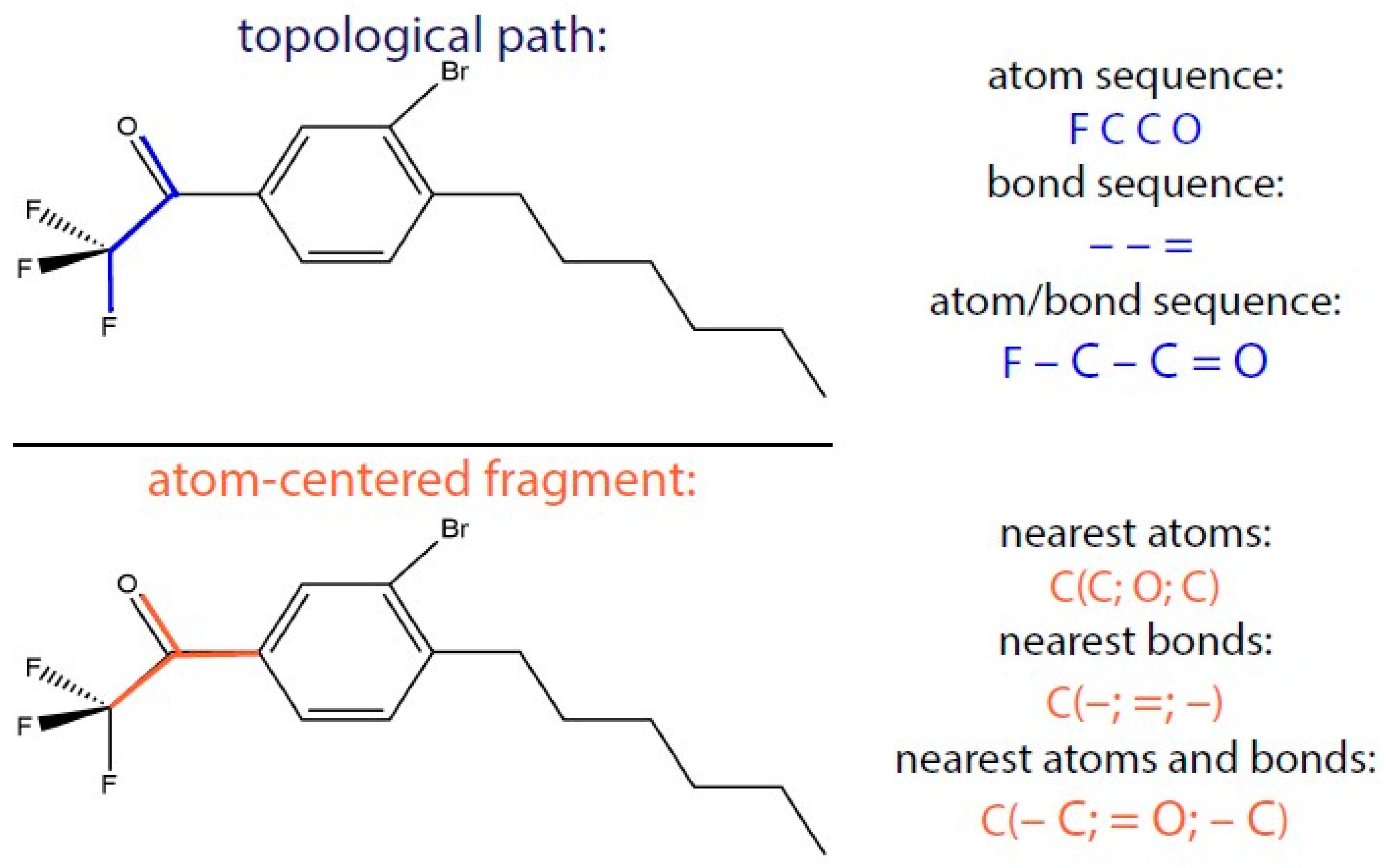
We used substructural molecular fragments (SMF) for encoding molecular structures in a matrix. A molecular structure can be described with this method by dividing a molecule into all possible fragments and writing the number of these fragments into the matrix. These SMFs were obtained by using “ISIDA QSPR” software [
7]. There are two approaches for obtaining a molecule’s SMF in ISIDA: sequences of atoms and/or bonds (topological path) and selected (“augmented”) atom (atom-centered fragments) with its environment that can be atoms, bonds, or both (
Figure 1). In this work, atom and bond sequences were applied. A molecule was represented as a graph and its descriptors were, consequently, subgraphs.
Hence, ISIDA SMF descriptors are numbers of fragments (or subgraphs) in a molecule with each element of the descriptor associated with one of the detected possible fragments. Only the shortest paths from one atom to the other were used. It should be noted that the length of sequences is limited. The minimal and the maximal lengths are 2 and 15, respectively.
2.3. Projection on Latent Structures (PLS) Modelling
In order to relate molecular descriptors of ligands with selectivity coefficients of the corresponding sensors, we employed the PLS regression algorithm. PLS regression searches for a set of components are known as latent vectors that perform a synchronous decomposition of X and Y, with the clause that these components explain, as much as possible, the covariance between X and Y. Data matrix size was 40 × 1855, where 40 is the number of samples (ligands) and 1855 is the number of descriptors.
3. Results and Discussion
The molecular descriptors obtained for the chosen ligands were calculated with ISIDA QSPR software [
7]. The 40 ionophore structures and their properties, specifically substructural molecular fragments (SMF) and the permittivity of membranes, were used as descriptors. The PLS model relating the descriptors with selectivity was evaluated according to the following parameters: root mean square error (RMSE) and squared determination coefficient (R
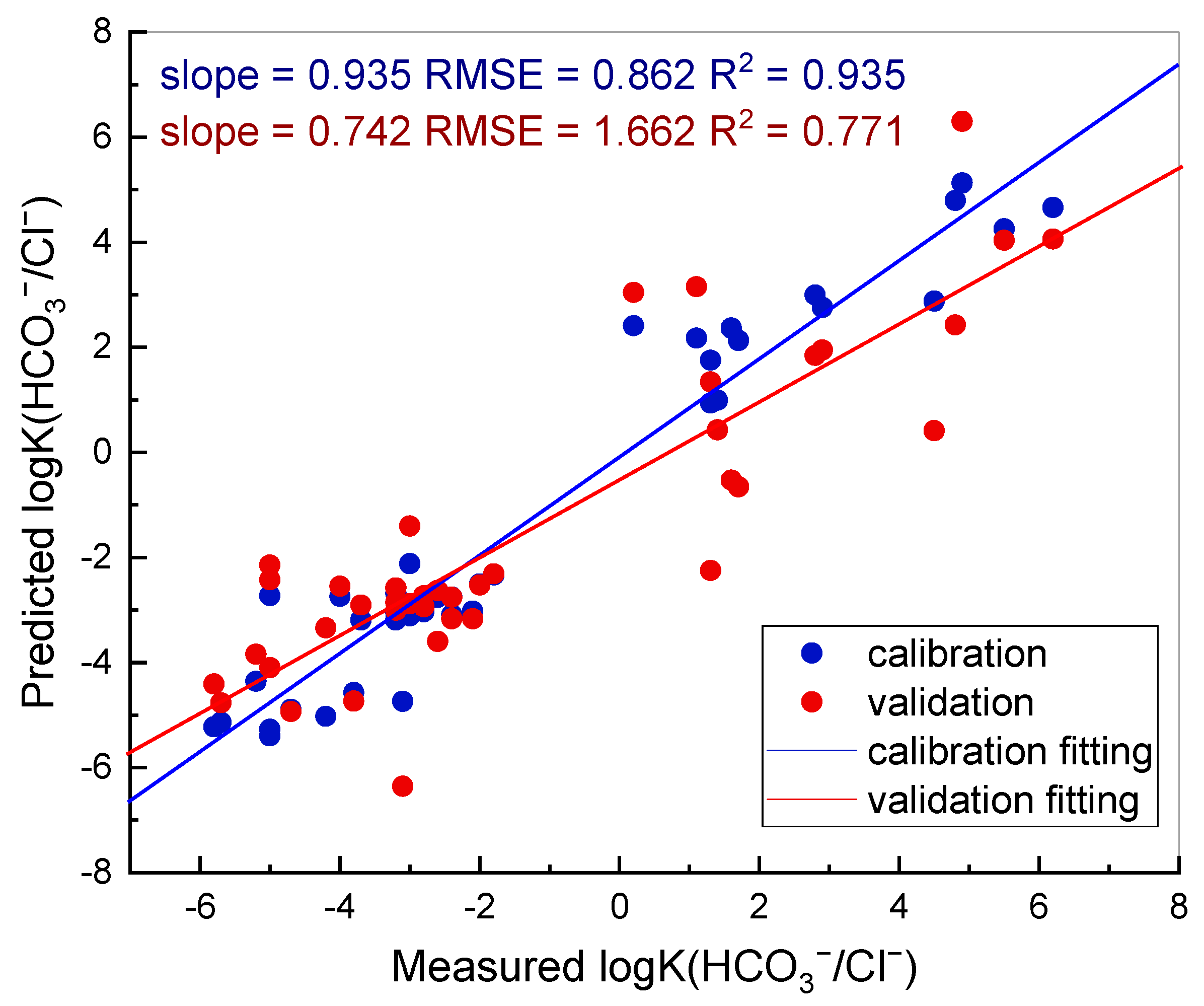
2). The results of QSPR modelling are shown in
Figure 2. Each point in the graph corresponds to an item in the database, whereas straight lines represent the resulting models. Blue and red colours correspond to training and test samples, respectively.
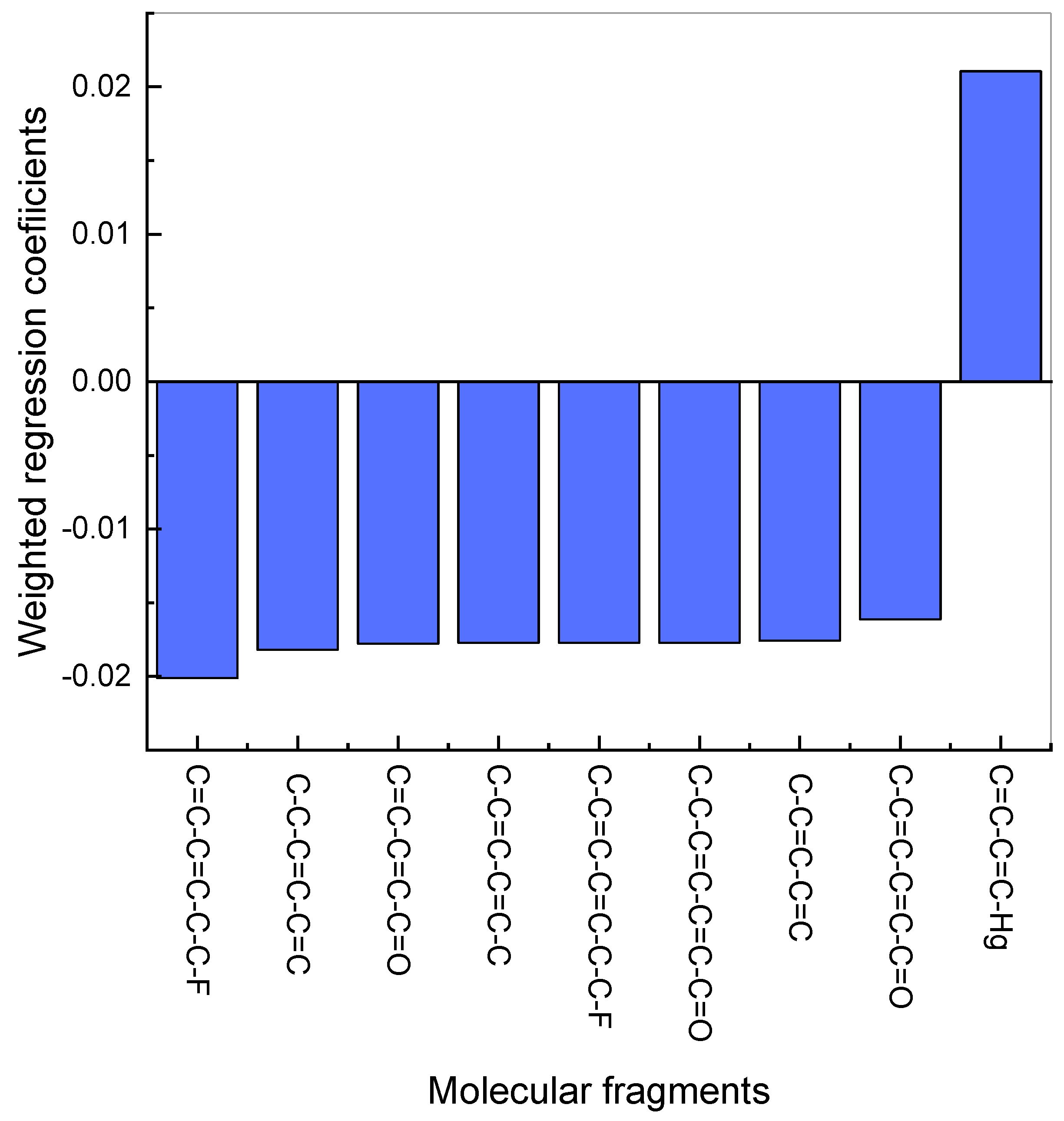
It can be seen that the derived model allows for a semi-quantitative estimation of the selectivity coefficients, based on the ligand structure. As far as each compound is an individual molecule, which consists of a variety of molecular fragments, regression coefficients allow evaluating the contribution of each fragment (represented as an independent variable in the matrix) in the selectivity of the sensors. The largest coefficients signify the variables (in this case, SMF, which are the encoded fragments of molecules) with the most important impact. The fragments with the largest contribution in the absolute value of the selectivity logK (HCO
3−/Cl
−) of potentiometric membrane sensors are shown in
Figure 3.
As follows from the graph, the fragment C=C-C=C-C-C-F makes the largest negative contribution, and it is part of a longer fragment C-C=C-C=C-C-C-F with a smaller contribution. The shortest fragment with negative contribution C-C=C-C=C is included in the remaining fragments with the largest negative contribution. The fragment with positive contribution C=C-C=C-Hg contains mercury in its composition. These observations provide valuable information for the further design of the ligands with required selectivity.
4. Conclusions
Despite some problems with anions’ ionophores that are described more fully in the introduction, we were able to collect the database that allowed making a QSPR model while satisfying RMSE and R2 for a relatively small amount of data. We found the fragments with the highest impact in selectivity logK (HCO3−/Cl−) of potentiometric membrane sensors, and we believe that this will help in the future search for the new ionophores. It appears that semi-quantitative prediction of sensor selectivity is possible based on the ligand structure.
Author Contributions
Conceptualization, D.K.; methodology, D.K.; formal analysis, D.K. and N.V.; investigation, N.V.; writing—original draft preparation, N.V.; writing—review and editing, D.K. and J.A.; visualization, N.V.; supervision, D.K.; funding acquisition, J.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by RFBR, grant number 20-33-70272.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kapetanovic, I.M. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chem.-Biol. Interact. 2008, 171, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruoff, R.; Tse, D.; Malhotra, R.; Lorents, D. Solubility of C60 in a Variety of Solvents. J. Phys. Chem. 1993, 97, 3379–3383. [Google Scholar] [CrossRef]
- Cruz, V.; Ramos, J.; Munoz-Escalona, A.; Lafuente, P.; Pena, B.; Martinez-Salazar, J. 3D-QSAR analysis of metallocene-based catalysts used in ethylene polymerization. Polymer 2004, 45, 2061–2072. [Google Scholar] [CrossRef]
- Trohalaki, S.; Pachter, R.; Drake, G.W.; Hawkins, T. Quantitative Structure-Property Relationships for Melting Points and Densities of Ionic Liquids. Energy Fuels 2005, 19, 279–284. [Google Scholar] [CrossRef]
- Martynko, E.; Solov’Ev, V.; Varnek, A.; Legin, A.; Kirsanov, D. QSPR Modeling of Potentiometric Mg2+/Ca2+ Selectivity for PVC-plasticized Sensor Membranes. Electroanalysis 2020, 32, 792–798. [Google Scholar] [CrossRef]
- Umezawa, Y.; Umezawa, K.; Bühlmann, P.; Hamada, N.; Aoki, H.; Nakanishi, J.; Sato, M.; Xiao, K.P.; Nishimura, Y. Potentiometric selectivity coefficients of ion-selective electrodes. Part II. Inorganic anions (IUPAC Technical Report). Pure Appl. Chem. 2002, 74, 923–994. [Google Scholar] [CrossRef] [Green Version]
- Varnek, A. ISIDA (In Silico Design and Data Analysis) Program. Available online: http://infochim.u-strasbg.fr/recherche/isida/index.php (accessed on 7 June 2012).
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).