1′-Homocarbocyclic Nucleoside Analogs with an Optically Active Substituted Bicyclo[2.2.1]Heptane Scaffold †
Abstract
:1. Introduction
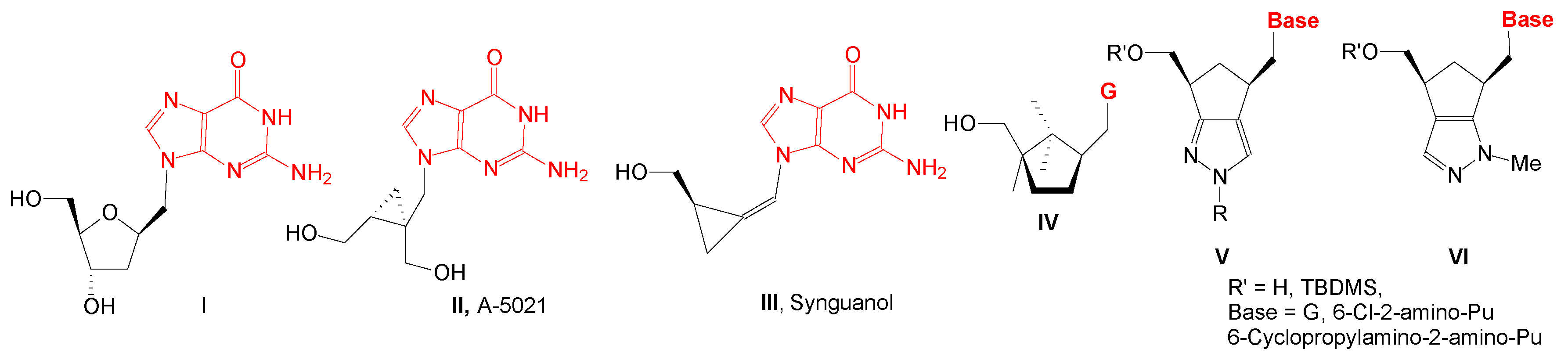
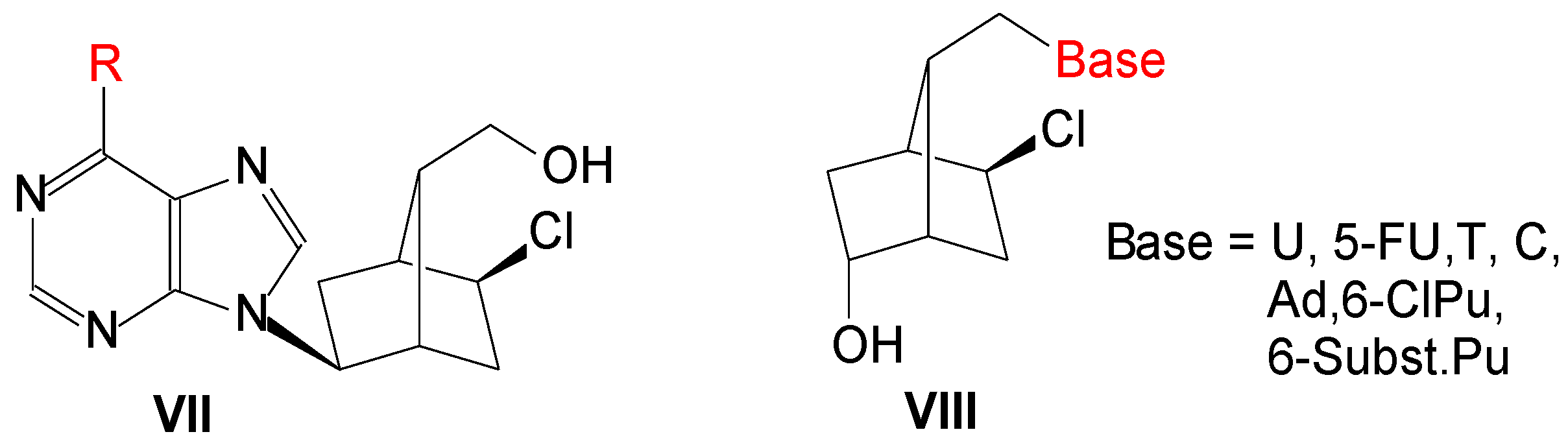
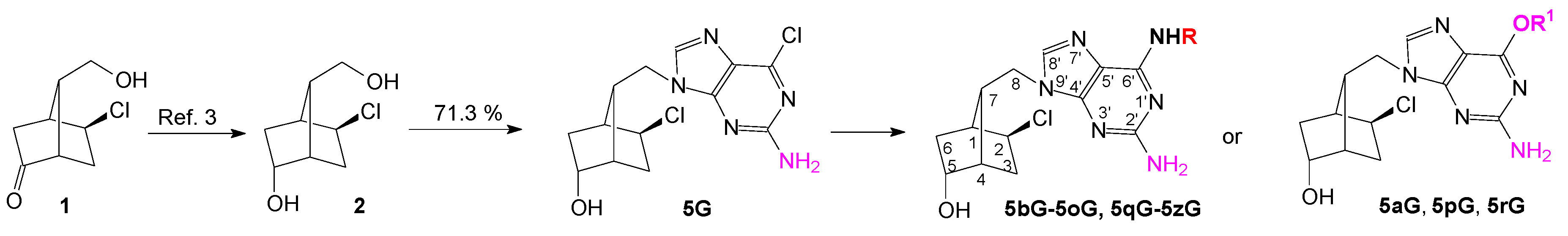
2. Results
3. Patents
Author Contributions
Funding
Conflicts of Interest
References
- Balzarini, J.; Naesens, L.; De Clercq, E. New antivirals—Mechanism of action and resistance development. Curr. Opin. Microbiol. 1998, 1, 535–546. [Google Scholar] [CrossRef]
- De Clerck, E.; Neyts, J. Antiviral Agents Acting as DNA or RNA Chain Terminator. In Handbook of Experimental Pharmacology 189; Kräusslich, H.-G., Bartenschlager, R., Eds.; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008; pp. 54–60. [Google Scholar]
- Tănase, C.; Drăghici, C.; Cojocaru, A.; Galochkina, A.V.; Orshanskaya, J.R.; Zarubaev, V.V.; Shova, S.; Enache, C.; Maganu, M. New carbocyclic N6-substituted adenine and pyrimidine nucleoside analogues with a bicyclo[2.2.1]heptane fragment as sugar moiety; synthesis, antiviral, anticancer activity and X-ray crystallography. Bioorg. Med. Chem. 2015, 23, 6346–6354. [Google Scholar] [CrossRef] [PubMed]
- Tănase, C.; Drăghici, C.; Hanganu, A.; Pintilie, L.; Maganu, M.; Volobueva, A.; Sinegubova, E.; Zarubaev, V.; Neyts, J.; Jochman, D.; et al. New HSV-1 Anti-Viral 1′-Homocarbocyclic Nucleoside Analogs with an Optically Active Substituted Bicyclo[2.2.1]Heptane Fragment as A Glycoside Moiety. Molecules 2019, 24, 2446. [Google Scholar] [CrossRef] [PubMed]
- Tănase, C.; Pintilie, L.; Mihai, E. New 1′-homocarbonucleoside Analogs with a Constrained Bicyclo[2.2.1]heptane Fragment. Patent Request A/00316/30.05, 2019. [Google Scholar]
- Tanase, C.; Pintilie, L. New 1′-homocarbanucleoside Analogs with a Constrained bicyclo[2.2.0]heptane Fragment and 2-amino-6-substituted Purine as Nucleobase. Patent request A/00290/27.05, 2020. [Google Scholar]
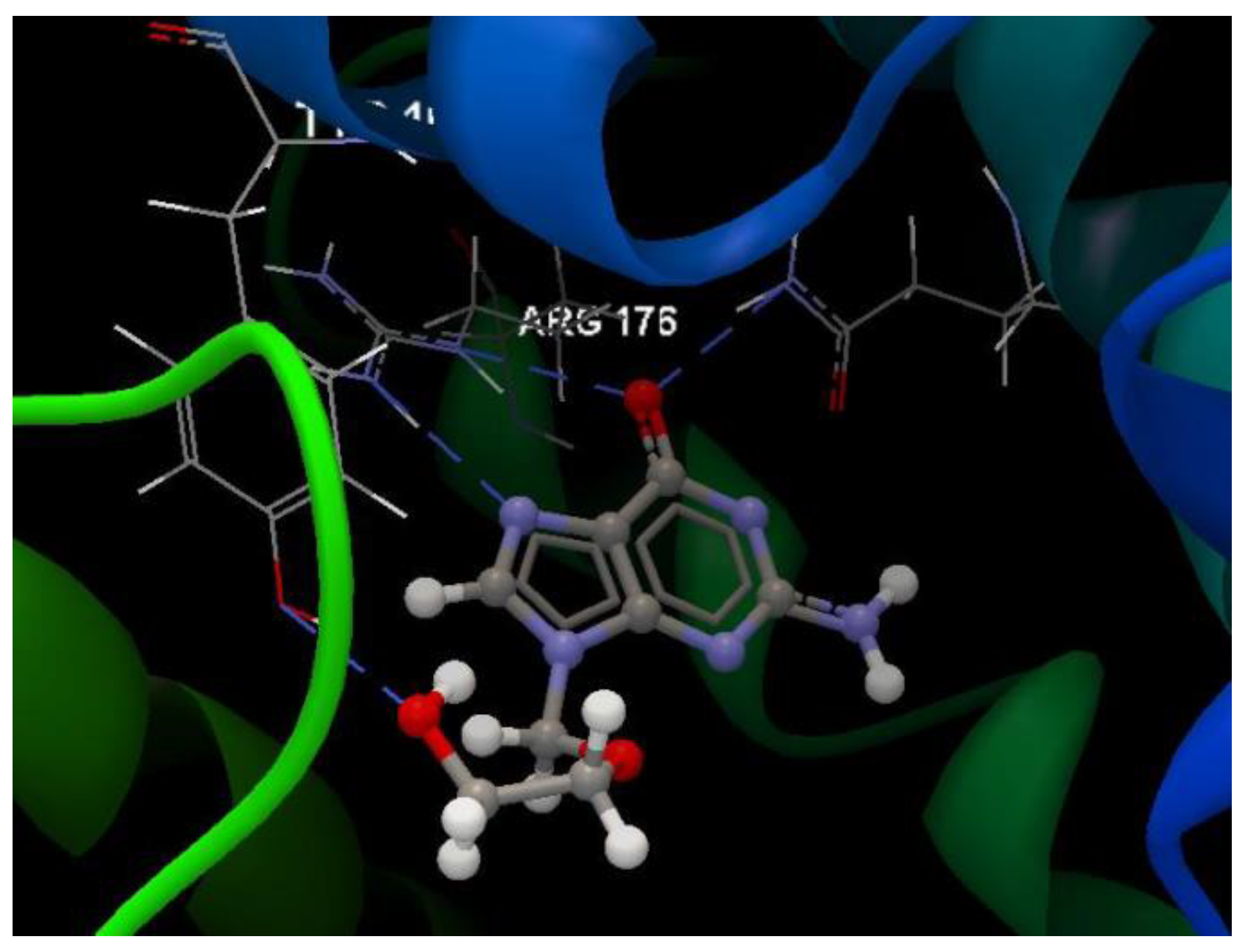
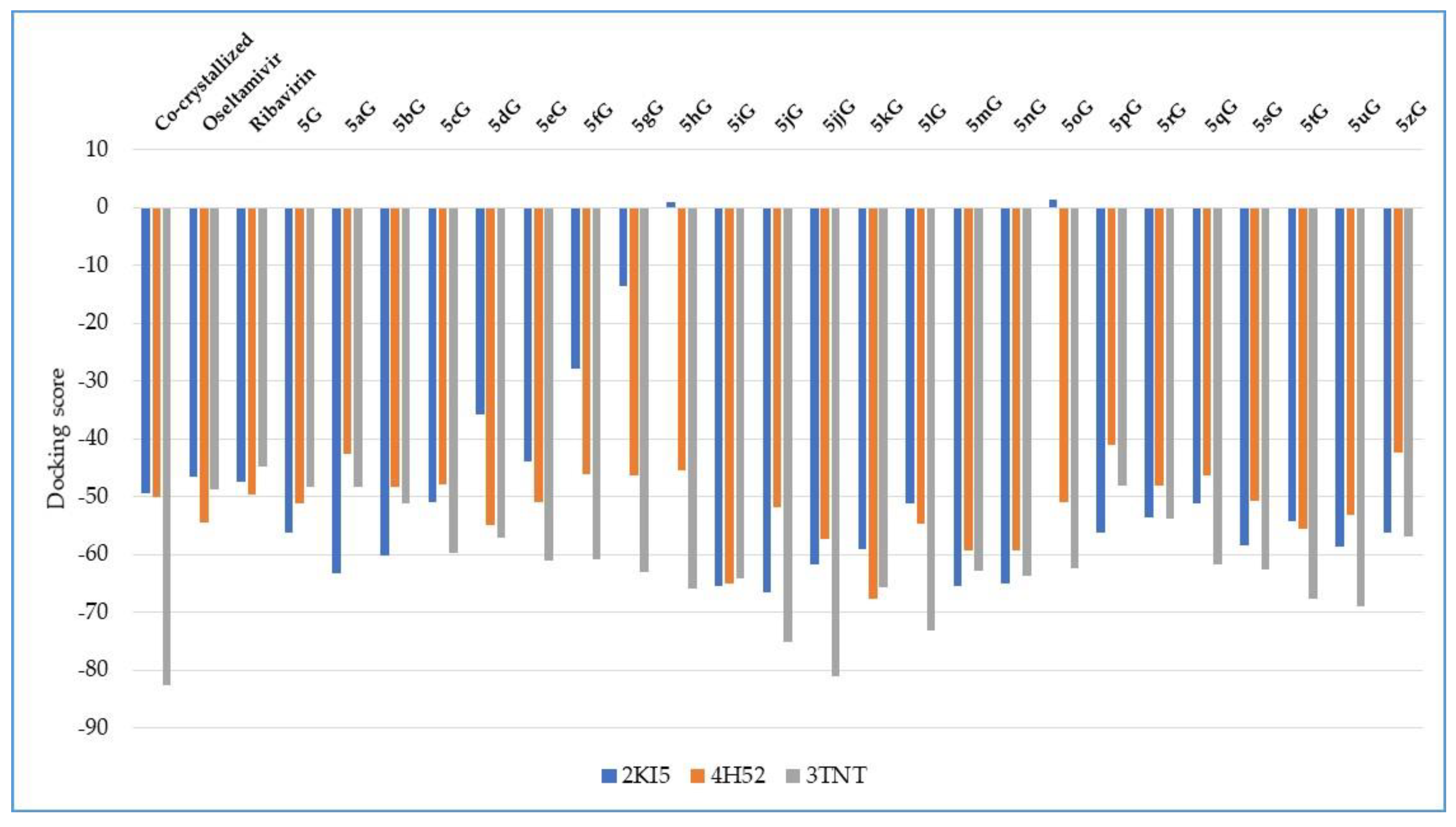
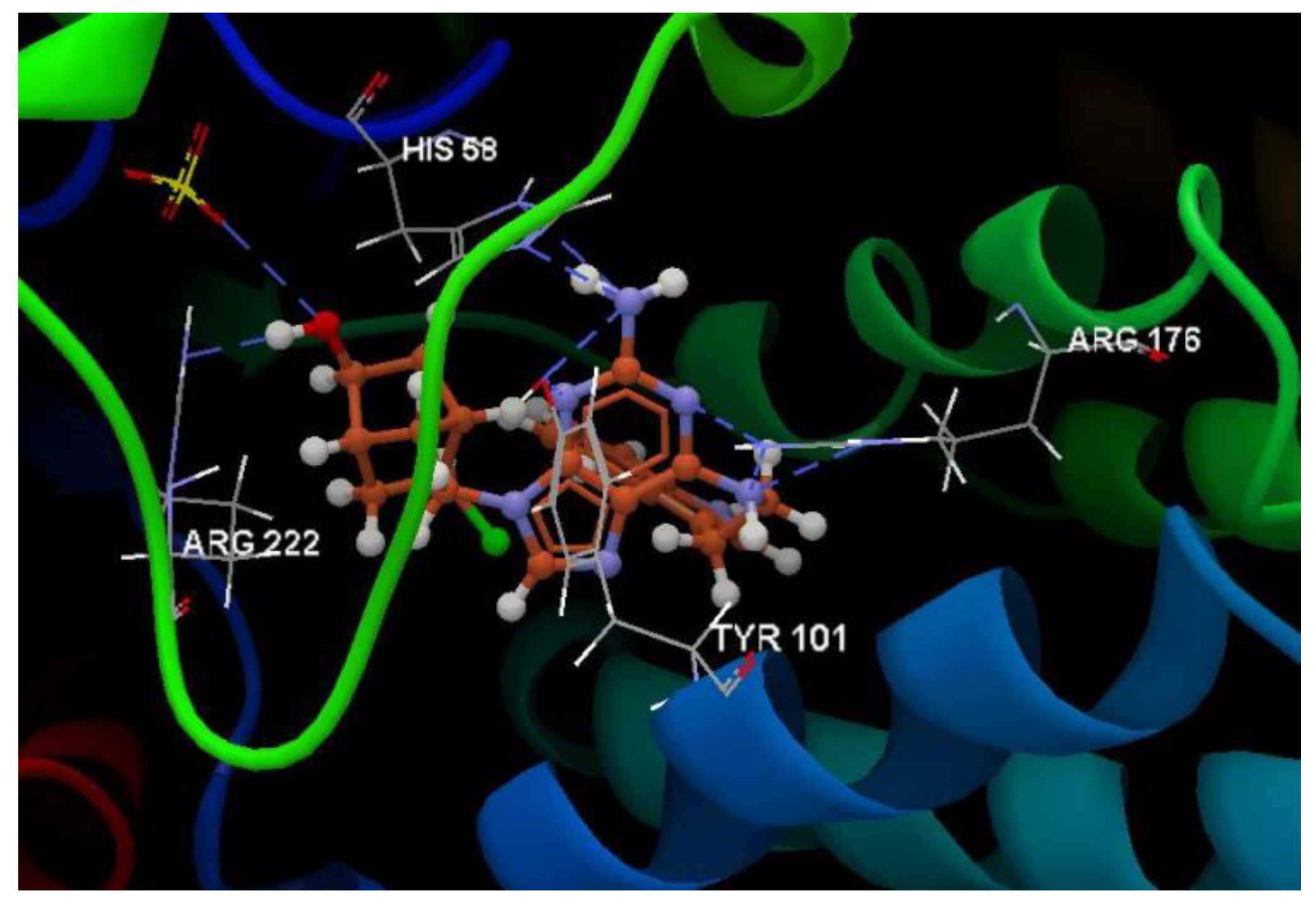


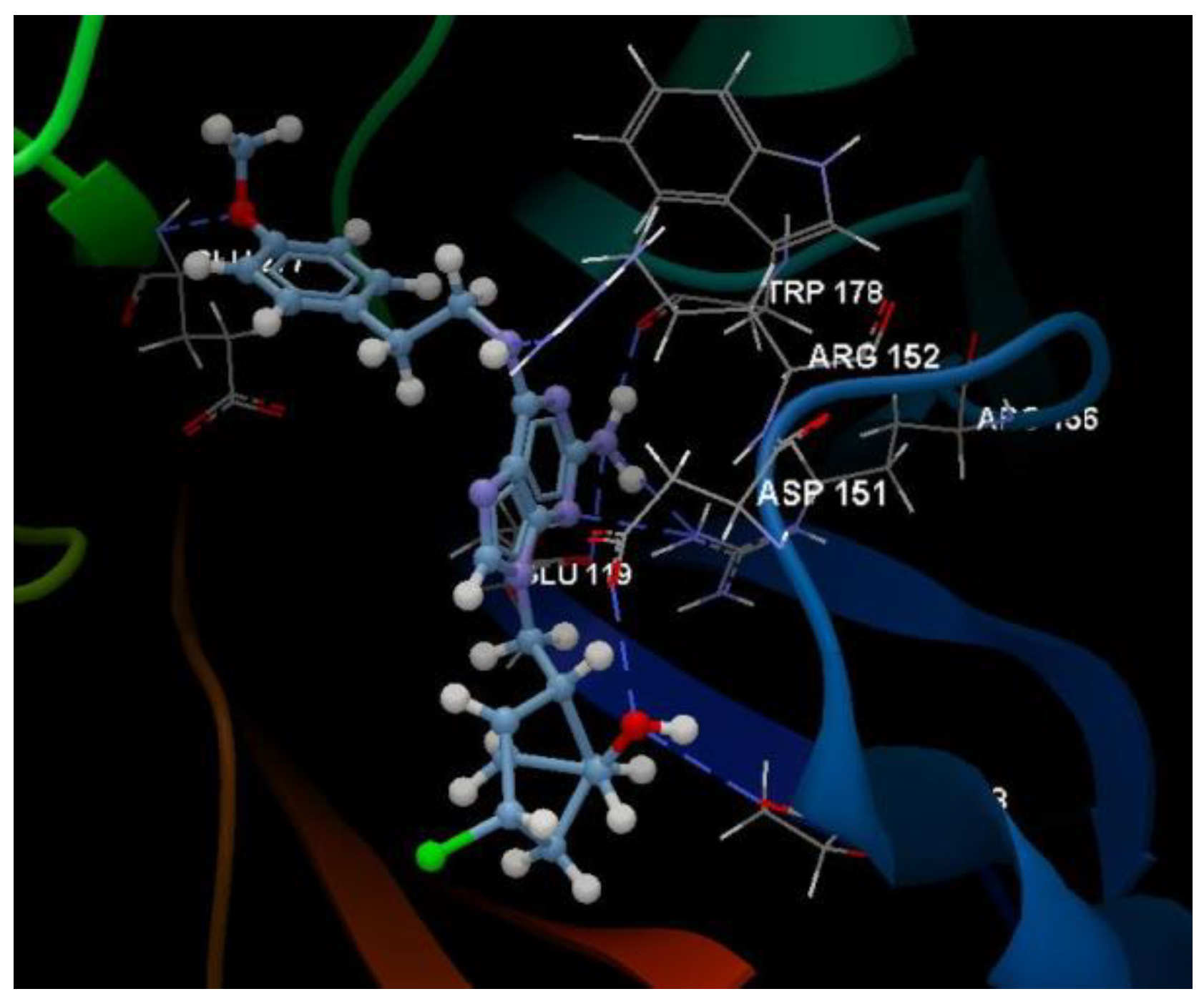
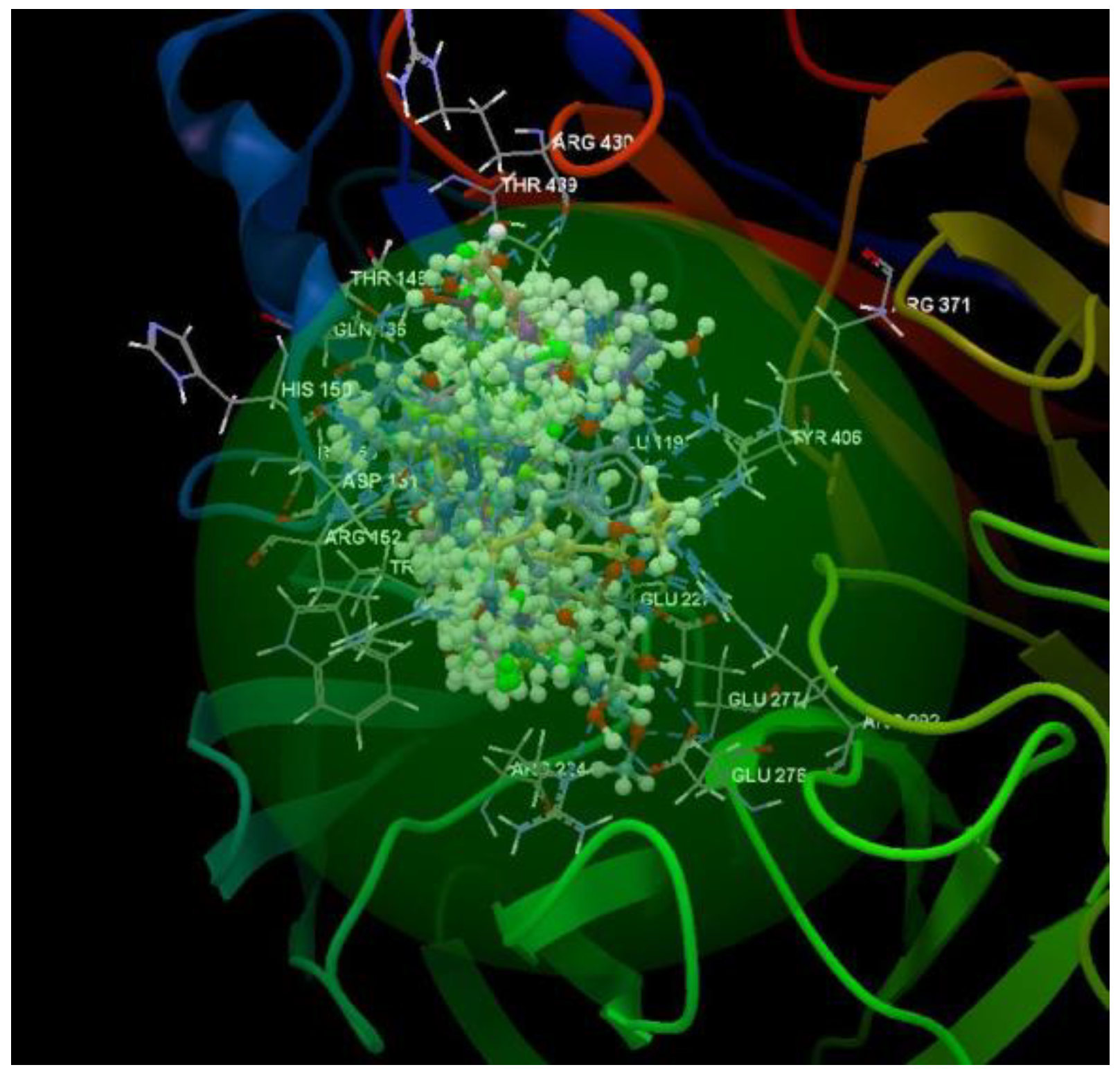
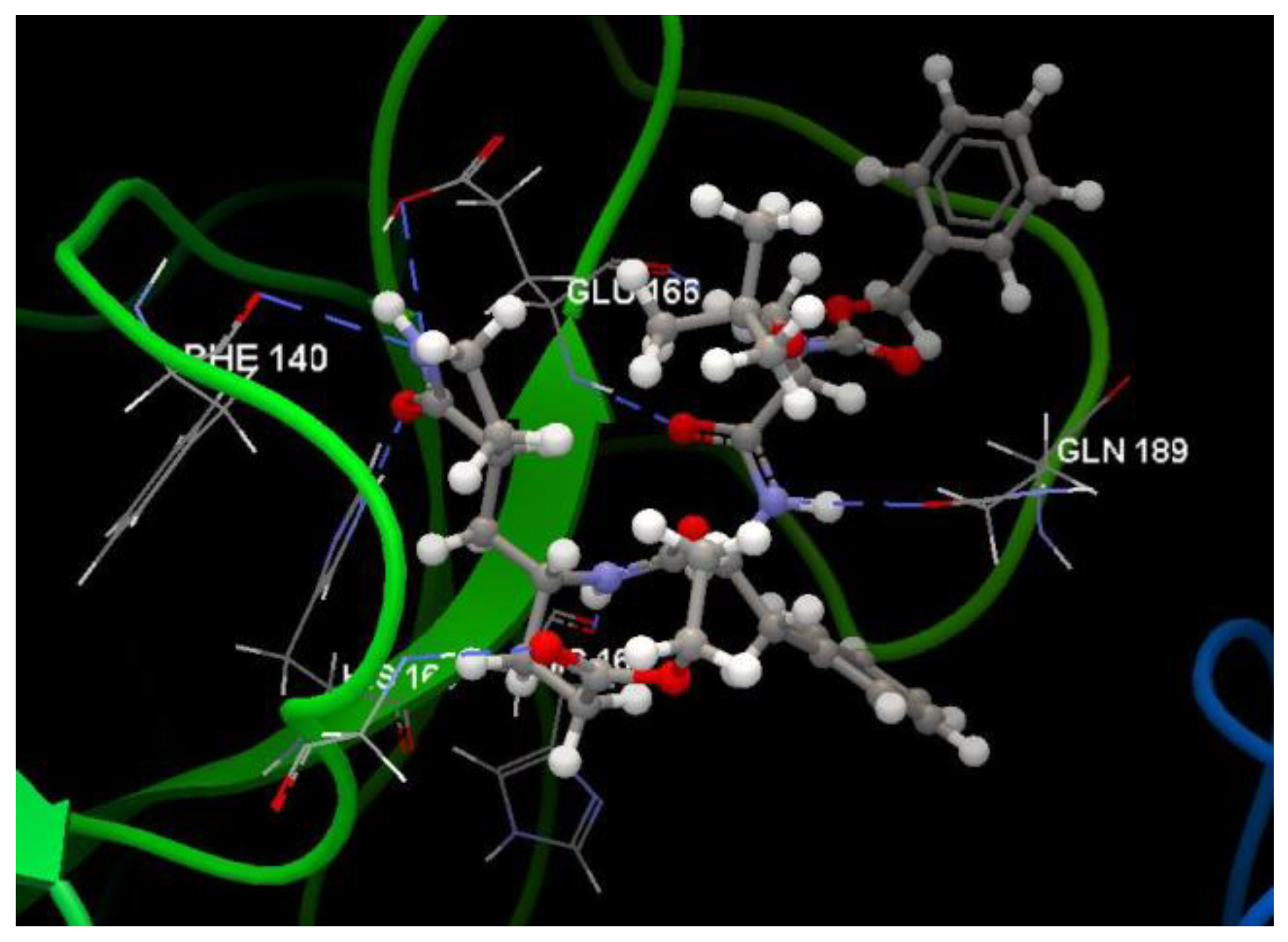
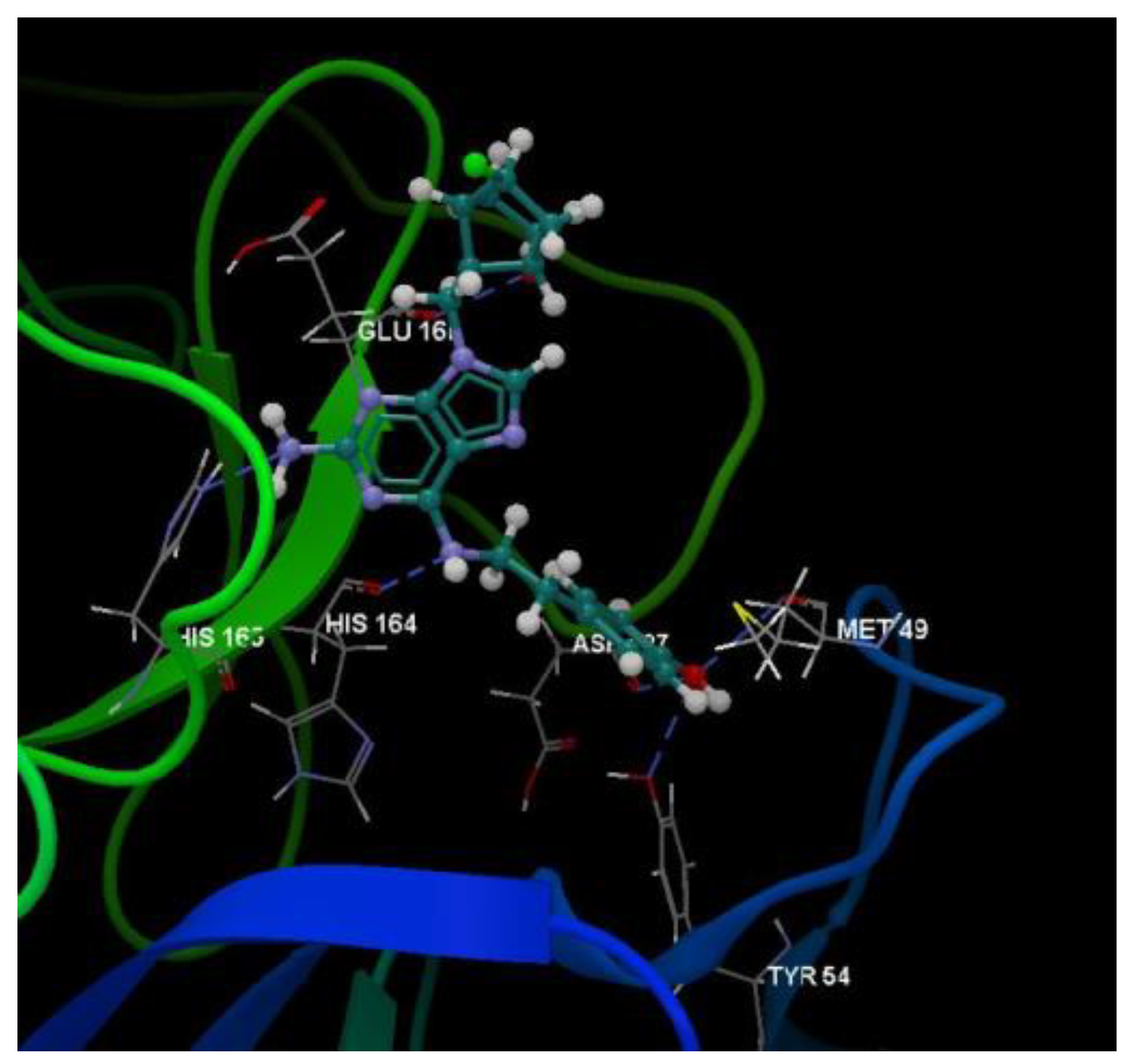
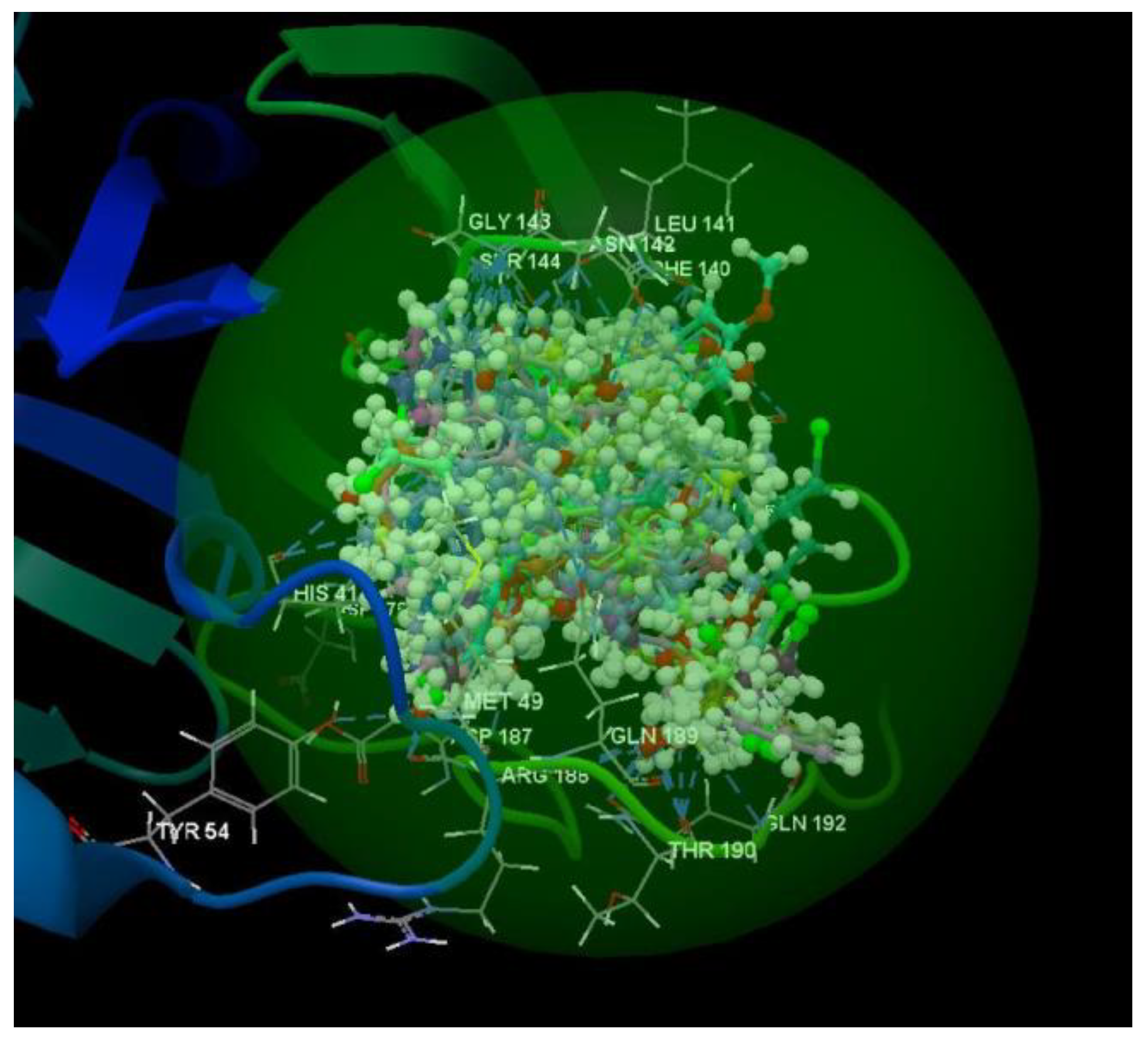
| Ligand | PDB ID: 2KI5 | PDB ID: 4H52 | PDB ID: 3TNT | |||
|---|---|---|---|---|---|---|
| Score | RMSD (Å) | Score | RMSD (Å) | Score | RMSD (Å) | |
| Co-crystallized | −49.29 | 0.71 | −49.94 | 0.21 | −82.61 | 2.41 |
| Oseltamivir | −46.52 | 0.02 | −54.44 | 0.62 | −48.75 | 1.96 |
| Ribavirin | −47.48 | 0.009 | −49.66 | 1.02 | −44.73 | 0.47 |
| 5G | −56.24 | 0.02 | −51.08 | 0.27 | −48.31 | 0.02 |
| 5aG | −63.14 | 0.01 | −42.49 | 0.01 | −48.31 | 0.04 |
| 5bG | −60.18 | 0.01 | −48.23 | 0.19 | −51.18 | 0.14 |
| 5cG | −50.98 | 0.05 | −47.88 | 0.87 | −59.63 | 0.28 |
| 5dG | −35.83 | 0.03 | −54.81 | 0.72 | −57.15 | 0.15 |
| 5eG | −43.80 | 0.23 | −50.95 | 1.04 | −60.92 | 0.35 |
| 5fG | −27.82 | 0.02 | −46.04 | 0.28 | −60.85 | 0.08 |
| 5gG | −13.47 | 0.18 | −46.27 | 0.10 | −62.98 | 0.50 |
| 5hG | +1.06 | 0.03 | −45.50 | 1.72 | −65.84 | 0.04 |
| 5iG | −65.51 | 0.19 | −64.95 | 0.28 | −64.07 | 1.46 |
| 5jG | −66.42 | 0.55 | −51.86 | 0.79 | −75.00 | 0.77 |
| 5jjG | −61.71 | 0.04 | −57.33 | 1.37 | −81.11 | 1.90 |
| 5kG | −59.01 | 0.60 | −67.57 | 0.58 | −65.73 | 0.55 |
| 5lG | −51.11 | 1.16 | −54.69 | 1.14 | −73.07 | 0.29 |
| 5mG | −65.46 | 0.45 | −59.33 | 0.78 | −62.74 | 1.21 |
| 5nG | −65.07 | 0.05 | −59.18 | 0.96 | −63.65 | 0.50 |
| 5oG | +1.37 | 0.04 | −50.88 | 0.19 | −62.38 | 0.45 |
| 5pG | −56.25 | 0.007 | −41.01 | 0.25 | −48.16 | 0.02 |
| 5rG | −53.58 | 0.22 | −48.02 | 0.09 | −53.66 | 0.06 |
| 5qG | −51.17 | 0.06 | −46.27 | 0.10 | −61.64 | 0.22 |
| 5sG | −58.31 | 0.02 | −50.72 | 0.99 | −62.51 | 0.54 |
| 5tG | −54.19 | 0.04 | −55.52 | 0.80 | −67.67 | 0.75 |
| 5uG | −58.66 | 0.13 | −53.11 | 0.03 | −68.96 | 0.42 |
| 5zG | −56.23 | 0.07 | −42.23 | 0.26 | −56.91 | 0.16 |
| Compound | Atoms | Weight锦(Daltons) | Flexible Bonds | Lipinski Violations | Hydrogen Donors | Hydrogen Acceptors | Log P |
|---|---|---|---|---|---|---|---|
| AC2 A * | 26 | 224.20 | 4 | 0 | 3 | 8 | 0.50 |
| FSI A 508 ** | 38 | 310.25 | 5 | 0 | 5 | 9 | −1.72 |
| G85A 501 *** | 95 | 652.78 | 20 | 2 | 4 | 12 | 3.69 |
| Oseltamivir | 50 | 312.40 | 8 | 0 | 3 | 6 | 1.70 |
| Ribavirin | 29 | 244.20 | 3 | 0 | 5 | 9 | −3.05 |
| 5G | 36 | 328.20 | 2 | 0 | 3 | 6 | 1.67 |
| 5aG | 37 | 309.75 | 2 | 0 | 4 | 7 | 0.69 |
| 5bG | 38 | 308.77 | 2 | 0 | 5 | 7 | 0.36 |
| 5cG | 45 | 348.83 | 4 | 0 | 4 | 7 | 1.58 |
| 5dG | 51 | 376.88 | 4 | 0 | 4 | 7 | 2.30 |
| 5eG | 54 | 390.91 | 4 | 0 | 4 | 7 | 2.84 |
| 5fG | 49 | 378.86 | 3 | 0 | 3 | 8 | 0.79 |
| 5gG | 53 | 391.90 | 3 | 0 | 3 | 8 | 0.98 |
| 5hG | 56 | 405.92 | 4 | 0 | 3 | 8 | 1.34 |
| 5iG | 54 | 412.92 | 6 | 0 | 4 | 7 | 2.98 |
| 5jG | 58 | 451.95 | 6 | 0 | 5 | 8 | 3.11 |
| 5jJG | 55 | 428.92 | 6 | 0 | 5 | 8 | 2.63 |
| 5kG | 58 | 442.94 | 7 | 0 | 4 | 8 | 2.95 |
| 5lG | 56 | 444.91 | 6 | 1 | 6 | 9 | 2.27 |
| 5mG | 54 | 416.91 | 7 | 0 | 4 | 9 | 1.10 |
| 5nG | 56 | 405.92 | 6 | 0 | 4 | 8 | 1.55 |
| 5oG | 58 | 420.94 | 5 | 0 | 5 | 9 | 0.04 |
| 5pG | 40 | 323.78 | 3 | 0 | 3 | 7 | 1.01 |
| 5rG | 43 | 337.80 | 4 | 0 | 3 | 7 | 1.38 |
| 5qG | 58 | 442.94 | 7 | 0 | 5 | 8 | 2.36 |
| 5sG | 50 | 399.88 | 5 | 0 | 4 | 8 | 1.49 |
| 5tG | 50 | 399.88 | 5 | 0 | 4 | 8 | 1.45 |
| 5uG | 50 | 399.88 | 5 | 0 | 4 | 8 | 1.45 |
| 5zG | 44 | 336.82 | 3 | 0 | 3 | 7 | 1.17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tănase, C.I.; Drăghici, C.; Hanganu, A.; Pintilie, L.; Maganu, M.; Zarubaev, V.V.; Volobueva, A.; Sinegubova, E. 1′-Homocarbocyclic Nucleoside Analogs with an Optically Active Substituted Bicyclo[2.2.1]Heptane Scaffold. Chem. Proc. 2021, 3, 16. https://doi.org/10.3390/ecsoc-24-08367
Tănase CI, Drăghici C, Hanganu A, Pintilie L, Maganu M, Zarubaev VV, Volobueva A, Sinegubova E. 1′-Homocarbocyclic Nucleoside Analogs with an Optically Active Substituted Bicyclo[2.2.1]Heptane Scaffold. Chemistry Proceedings. 2021; 3(1):16. https://doi.org/10.3390/ecsoc-24-08367
Chicago/Turabian StyleTănase, Constantin I., Constantin Drăghici, Anamaria Hanganu, Lucia Pintilie, Maria Maganu, Vladimir V. Zarubaev, Alexandrina Volobueva, and Ekaterina Sinegubova. 2021. "1′-Homocarbocyclic Nucleoside Analogs with an Optically Active Substituted Bicyclo[2.2.1]Heptane Scaffold" Chemistry Proceedings 3, no. 1: 16. https://doi.org/10.3390/ecsoc-24-08367
APA StyleTănase, C. I., Drăghici, C., Hanganu, A., Pintilie, L., Maganu, M., Zarubaev, V. V., Volobueva, A., & Sinegubova, E. (2021). 1′-Homocarbocyclic Nucleoside Analogs with an Optically Active Substituted Bicyclo[2.2.1]Heptane Scaffold. Chemistry Proceedings, 3(1), 16. https://doi.org/10.3390/ecsoc-24-08367







