Exploring Green Tea Polyphenols Against Penicillin-Binding Proteins (PBPs) as Prospective Targets for Peptic Ulcer Treatment: In Silico Analysis †
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Docking
2.2. In Silico Drug-Likeness and ADMET Analysis
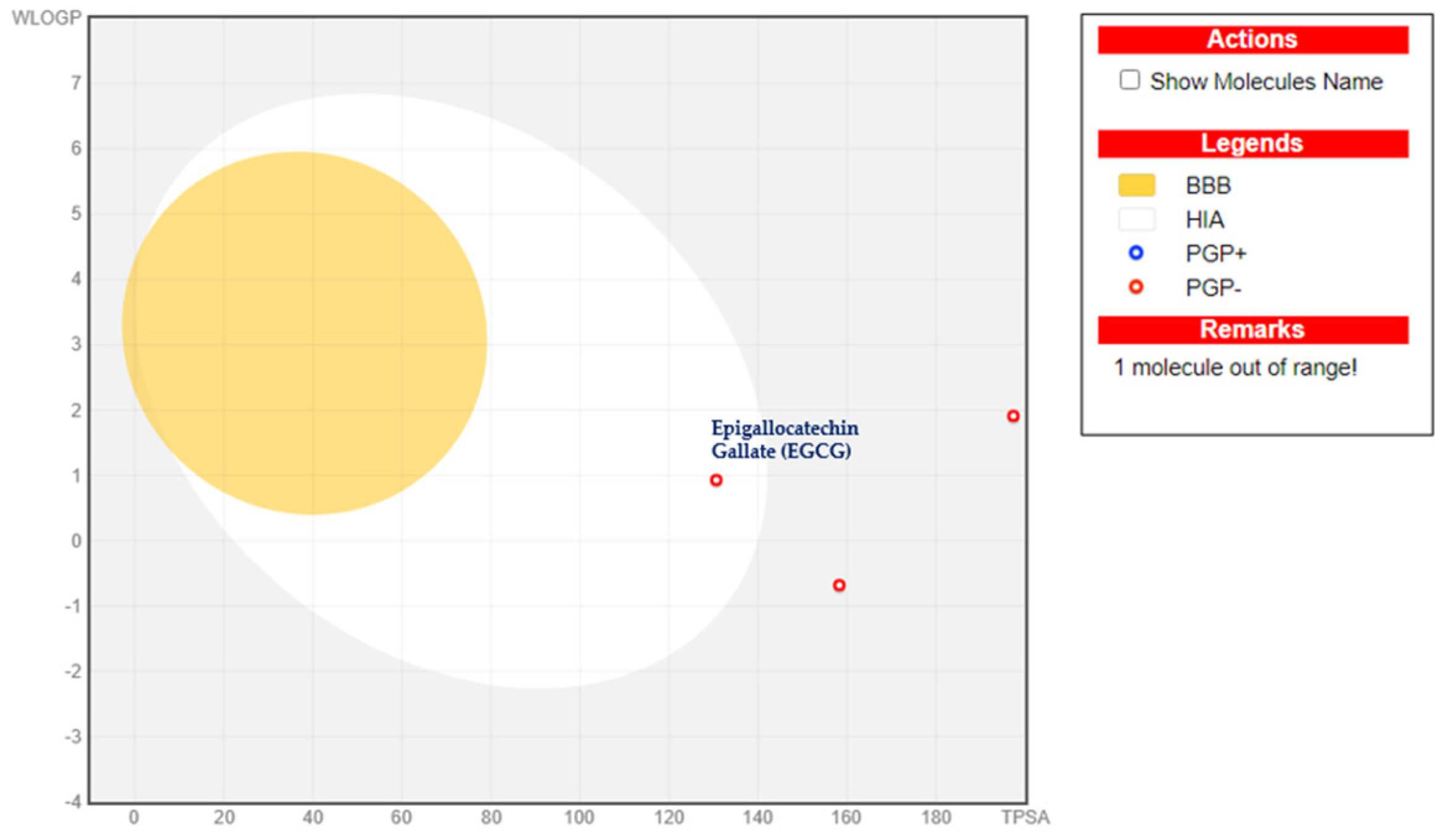
2.3. BOILED-Egg Model Analysis
3. Results and Discussion
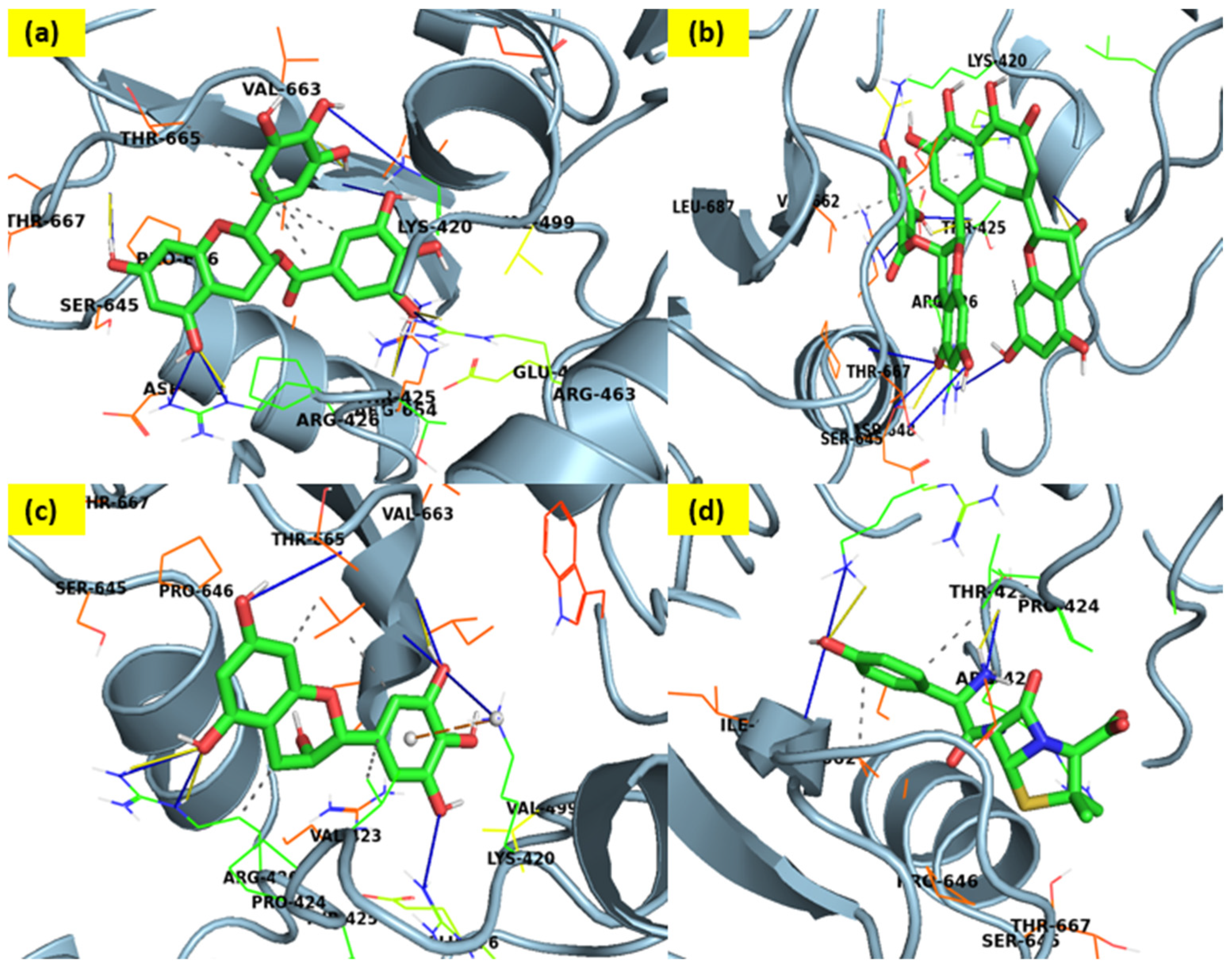
3.1. Docking Interaction Analyses
| Molecules | Binding Affinity (kcal/mol) | Molecules | Binding Affinity (kcal/mol) |
|---|---|---|---|
| Oolonghomobisflavan A | Theaflavic Acid | −7.21 | |
| Theasinensin D | −9.74 | Barrigenol R1 | |
| Theaflavin-3-gallate | −16.57 | Barringtogenol | −9.3 |
| Isotheaflavin | −14.1 | Camelliagenin | −6.41 |
| Epigallocatechin-3,5-Di-O-Gallate | −6.67 | Gallocatechin | −6.14 |
| Oolonghomobisflavan B | −12.5 | Catechin | −6.01 |
| Cis-3-Hexenol | −4.1 | Epicatechin | −5.28 |
| Epigallocatechin-3,4-Di-O-Gallate | −12.0 | Epiafzelechin | −4.07 |
| Vicenin 2 | −5.33 | Quercetin | −9.02 |
| Epicatechin-3,5-Di-O-Gallate | −11.4 | Cryptoxanthin | −9.11 |
| Rutin | −5.98 | Myricetin | −11.4 |
| Proanthocyanidin | −5.33 | Apigenin | −4.99 |
| Pheophytin | −4.22 | Nerolidol | −4.36 |
| Benzaldehyde | −4.8 | Kaempferol | −3.10 |
| Epitheaflavic Acid 3′-Gallate | −4.19 | Theanine | −2.90 |
| Epigallocatechin Gallate | −17.23 | Ascorbic Acid | −2.11 |
| Theasinensin E | −7.32 | Quinic Acid | −1.09 |
| Myricitrin | −11.4 | Succinic Acid | −1.7 |
| Theaflavin | −2.23 | Methyl Salicylate | −5.37 |
| Epicatechin Gallate | −9.1 | Theobromine | −5.21 |
| Kaempferitrin | −4.73 | Caffeine | −5.78 |
| Isoquercetin | −9.6 | Xanthine | −5.59 |
| Epiafzelechin 3-O-Gallate | −11.0 | Linalool Oxide | −5.88 |
| Pheophorbide | −7.34 | Phenylacetaldehyde | −5.71 |
| Epigallocatechin 3-O-P-Coumarate | −7.25 | Methylxanthine | −5.66 |
| Pheophorbide | −5.55 | Theophylline | −5.69 |
| Oxalic Acid | −5.03 | Geraniol | −5.31 |
| Cryptoxanthin | −5.21 | Hexanal | −5.36 |
| Isovitexin | −5.19 | Diphenylamine | −3.9 |
| Vitexin | −5.01 | Trans-2-Hexenal | −5.99 |
| Chlorogenic Acid | −4.09 | Linalool | −6.03 |
| Coumaroyl Quinic Acid | −7.02 | Phenylethanol | −6.07 |
| Epigallocatechin | −15.91 | Amoxicillin (Std.) | −10.93 |
| Sr. No. | Molecules | Docking Score (kcal/mol) | Residues with Contribution Energy |
|---|---|---|---|
| 1. | Amoxicillin (Std.) | −10.93 | Thr 526, Trp 374, Ser 337, Ser 395, Thr 550, Met 527, and Tyr 595 |
| 2. | Theaflavin-3-gallate | −16.57 | Trp374; Arg372; Phe570; Thr550; Ser548; Ser337; Asp373 |
| 3. | Epigallocatechin | −15.91 | Asn377; Trp374; Thr550; Phe450 |
| 4 | Epigallocatechin Gallate (EGCG) (Best docked) | −17.23 | Trp374; Gln552;Phe450;Gln452;Ser337; Phe570;Tyr568;Asn377;Arg372;Gly451;Gly336;Glu334;Ala551;Lys340;Ser395;Thr526;Phe570 |

3.2. The ADME Analysis and BOILED-Egg
| Properties | Theaflavin-3-Gallate | Epigallocatechin | Epigallocatechin Gallate (EGCG) * |
|---|---|---|---|
| CYP450 2C9 Substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 2D6 Substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 3A4 Substrate | Non-substrate | Non-substrate | Non-substrate |
| Human Ether-a-go-go-Related Gene Inhibition | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| AMES Toxicity | Non-AMES toxic | Non-AMES toxic | Non-AMES toxic |
| Carcinogens | None | None | None |
| Acute Oral Toxicity | IV | IV | IV |
| P-glycoprotein Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| Rat Acute Toxicity (LD50, mol/kg) | 2.6693 | 1.8700 | 2.6643 |
| Human Intestinal Absorption | + | + | + |
| AlogP | 3.19 | 1.25 | 2.23 |
| H-Bond Acceptor | 16 | 7 | 11 |
| H-Bond Donor | 11 | 6 | 8 |
| Tetrahymena pyriformis (pIGC50 (ug/L)) | 0.595 | 0.792 | 0.913 |
| Blood–Brain Barrier | - | - | - |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yadav, S.; Pandey, A.; Mali, S.N. From Lab to Nature: Recent Advancements in the Journey of Gastroprotective Agents from Medicinal Chemistry to Phytotherapy. Eur. J. Med. Chem. 2024, 272, 116436. [Google Scholar] [CrossRef] [PubMed]
- Sugano, K.; Tack, J.; Kuipers, E.J.; Graham, D.Y.; El-Omar, E.M.; Miura, S.; Haruma, K.; Asaka, M.; Uemura, N.; Malfertheiner, P. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 2015, 64, 1353–1367. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Valle, M.; Orellana-Palma, P.; Petzold, G. Plant-based polyphenols: Anti-Helicobacter pylori effect and improvement of gut microbiota. Antioxidants 2022, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Scortecci, K.C.; Boylan, F. A Review on Flavonoids as Anti-Helicobacter pylori Agents. Appl. Sci. 2025, 15, 3936. [Google Scholar] [CrossRef]
- Gerrits, M.M.; Schuijffel, D.; Van Zwet, A.A.; Kuipers, E.J.; Vandenbroucke-Grauls, C.M.J.E.; Kusters, J.G. Alterations in penicillin-binding protein 1A confer resistance to β-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 2002, 46, 2229–2233. [Google Scholar] [CrossRef] [PubMed]
- Makarewicz, M.; Drożdż, I.; Tarko, T.; Duda-Chodak, A. The Interactions between polyphenols and microorganisms, especially gut microbiota. Antioxidants 2021, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Filosa, S.; di Meo, F.; Crispi, S. Polyphenols-gut microbiota interplay and brain neuromodulation. Neural Regen. Res. 2018, 13, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Mali, S.N.; Thorat, B.R.; Gupta, D.R.; Pandey, A. Mini-Review of the Importance of Hydrazides and Their Derivatives—Synthesis and Biological Activity. Eng. Proc. 2021, 11, 21. [Google Scholar] [CrossRef]
- Kabier, M.; Gambacorta, N.; Trisciuzzi, D.; Kumar, S.; Nicolotti, O.; Mathew, B. MzDOCK: A free ready-to-use GUI-based pipeline for molecular docking simulations. J. Comput. Chem. 2024, 45, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayyappan, P.; Rao, J.; Ganpule, P.; Somani, R.; Mali, S.N. Exploring Green Tea Polyphenols Against Penicillin-Binding Proteins (PBPs) as Prospective Targets for Peptic Ulcer Treatment: In Silico Analysis. Chem. Proc. 2025, 18, 89. https://doi.org/10.3390/ecsoc-29-26744
Ayyappan P, Rao J, Ganpule P, Somani R, Mali SN. Exploring Green Tea Polyphenols Against Penicillin-Binding Proteins (PBPs) as Prospective Targets for Peptic Ulcer Treatment: In Silico Analysis. Chemistry Proceedings. 2025; 18(1):89. https://doi.org/10.3390/ecsoc-29-26744
Chicago/Turabian StyleAyyappan, Parasuram, Janavi Rao, Pratik Ganpule, Rakesh Somani, and Suraj N. Mali. 2025. "Exploring Green Tea Polyphenols Against Penicillin-Binding Proteins (PBPs) as Prospective Targets for Peptic Ulcer Treatment: In Silico Analysis" Chemistry Proceedings 18, no. 1: 89. https://doi.org/10.3390/ecsoc-29-26744
APA StyleAyyappan, P., Rao, J., Ganpule, P., Somani, R., & Mali, S. N. (2025). Exploring Green Tea Polyphenols Against Penicillin-Binding Proteins (PBPs) as Prospective Targets for Peptic Ulcer Treatment: In Silico Analysis. Chemistry Proceedings, 18(1), 89. https://doi.org/10.3390/ecsoc-29-26744





