Novel Chalcone Derivatives as Potent Lyn Tyrosine Kinase Inhibitors: A Promising In-Silico Approach for Targeted Therapy in Triple-Negative Breast Cancer †
Abstract
1. Introduction
2. Materials and Methods
2.1. Software, Hardware, and Databases
2.2. Protein Crystal Structure Retrieval
2.3. Creation of Library
2.4. Evaluation of Theoretical Oral Bioavailability and Toxicity
2.5. Protein Structure Preparation
2.6. Ligand Structure Preparation
2.7. Molecular Docking Analysis
3. Results
3.1. Summary of the Designed Chalcone Library
3.2. Predicted Oral Bioavailability and Toxicity
ADMET Analysis
3.3. Molecular Docking Results
3.3.1. Grid Point Generation
| Enzyme | Grid-Box Size | Center | ||||
|---|---|---|---|---|---|---|
| X | Y | Z | X | Y | Z | |
| Lyn-kinase | 48 | 44 | 60 | 19.669 | −9.388 | 23.998 |
3.3.2. Validation of Docking Procedures
| Enzyme | Crystal Structure Complex (Enzyme and Ligand) | Crystal Structure Complex with Re-Docked Ligand (Validation) |
|---|---|---|
| 2zva | 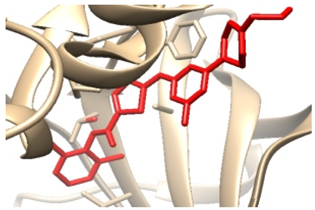 | 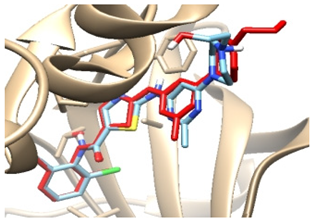 |
3.3.3. Binding Affinity of Ligands to Protease Enzymes
| Compound Name | SMILE | Dock Scores (kcal/mol) |
|---|---|---|
| Lig 0 | Dasatinib | −9.8 |
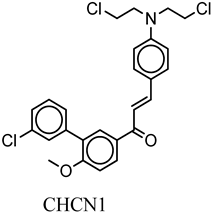
| CHCN1 | c1ccc(Cl)cc1c(c(cc2)OC)cc2C(=O)C=Cc3ccc(cc3)N(CCCl)CCCl | −8.6 |
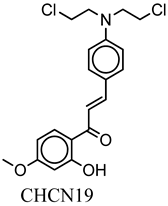
| CHCN19 | COc(cc1)cc(O)c1C(=O)C=Cc2ccc(cc2)N(CCCl)CCCl | −7.1 |
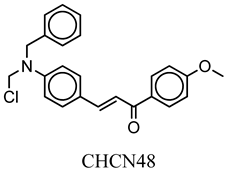
| CHCN48 | COc(cc1)ccc1C(=O)C=Cc2ccc(cc2)N(CCl)Cc3ccccc3 | −8.3 |
| CHCN333 | ClCCN(CCCl)c(cc1)ccc1C=CC(=O)c2cc(c(cc2)OC)Oc3ccccc3 | −8.1 |
| CHCN94 | COc(cc1)c(O)cc1C(=O)C=Cc2ccc(cc2)N(CCl)Cc3ccccc3 | −8.0 |
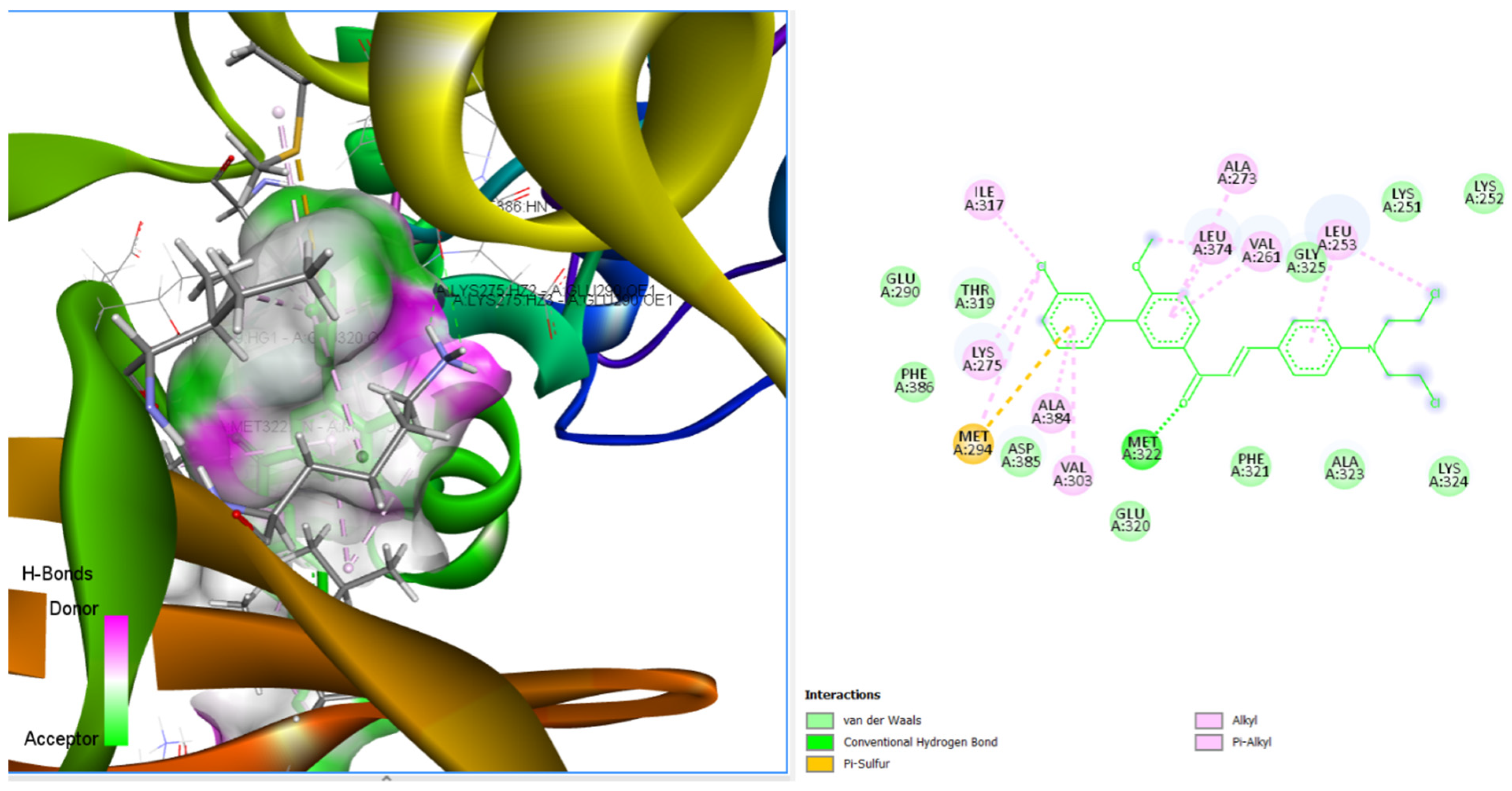
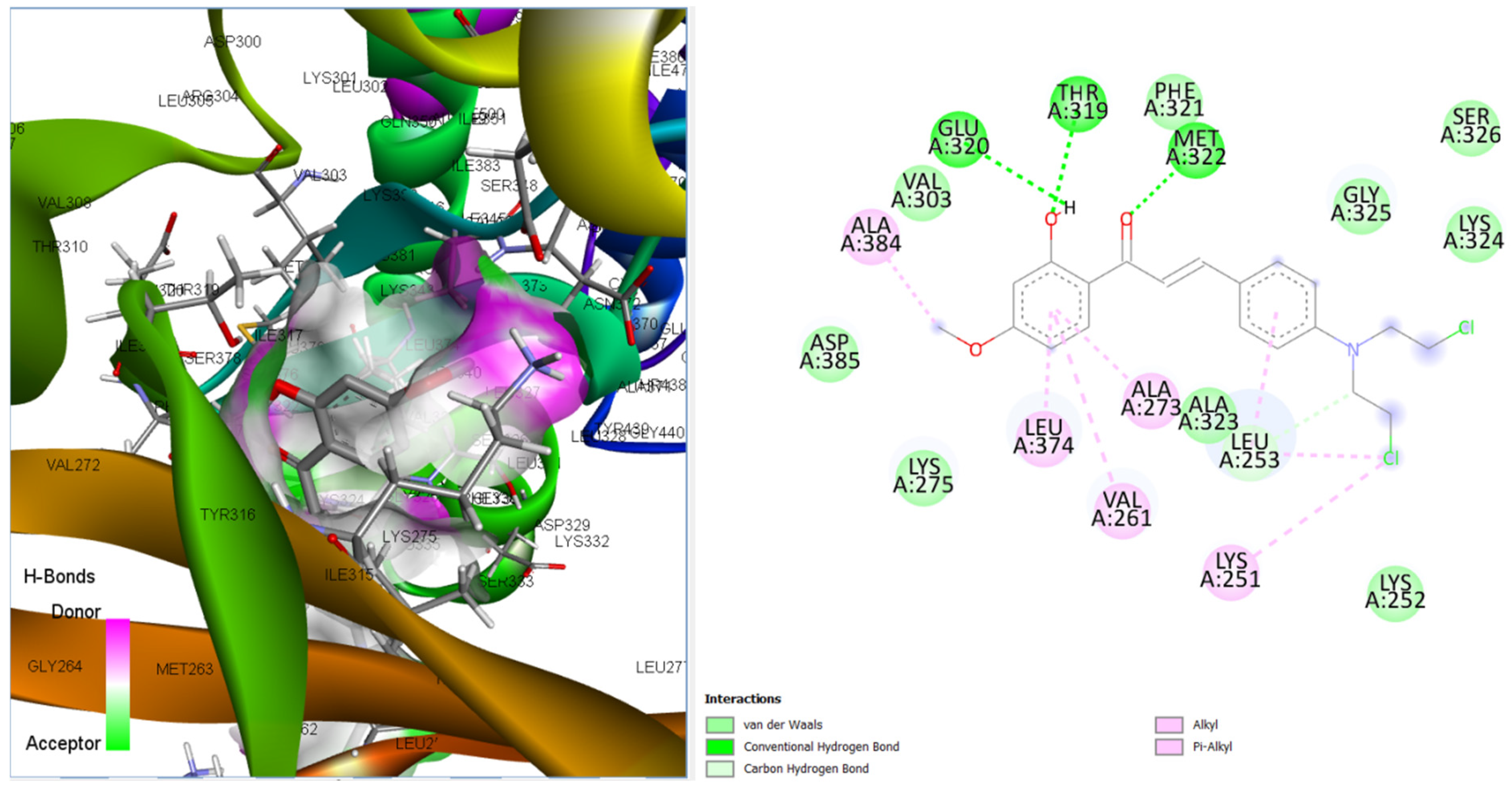
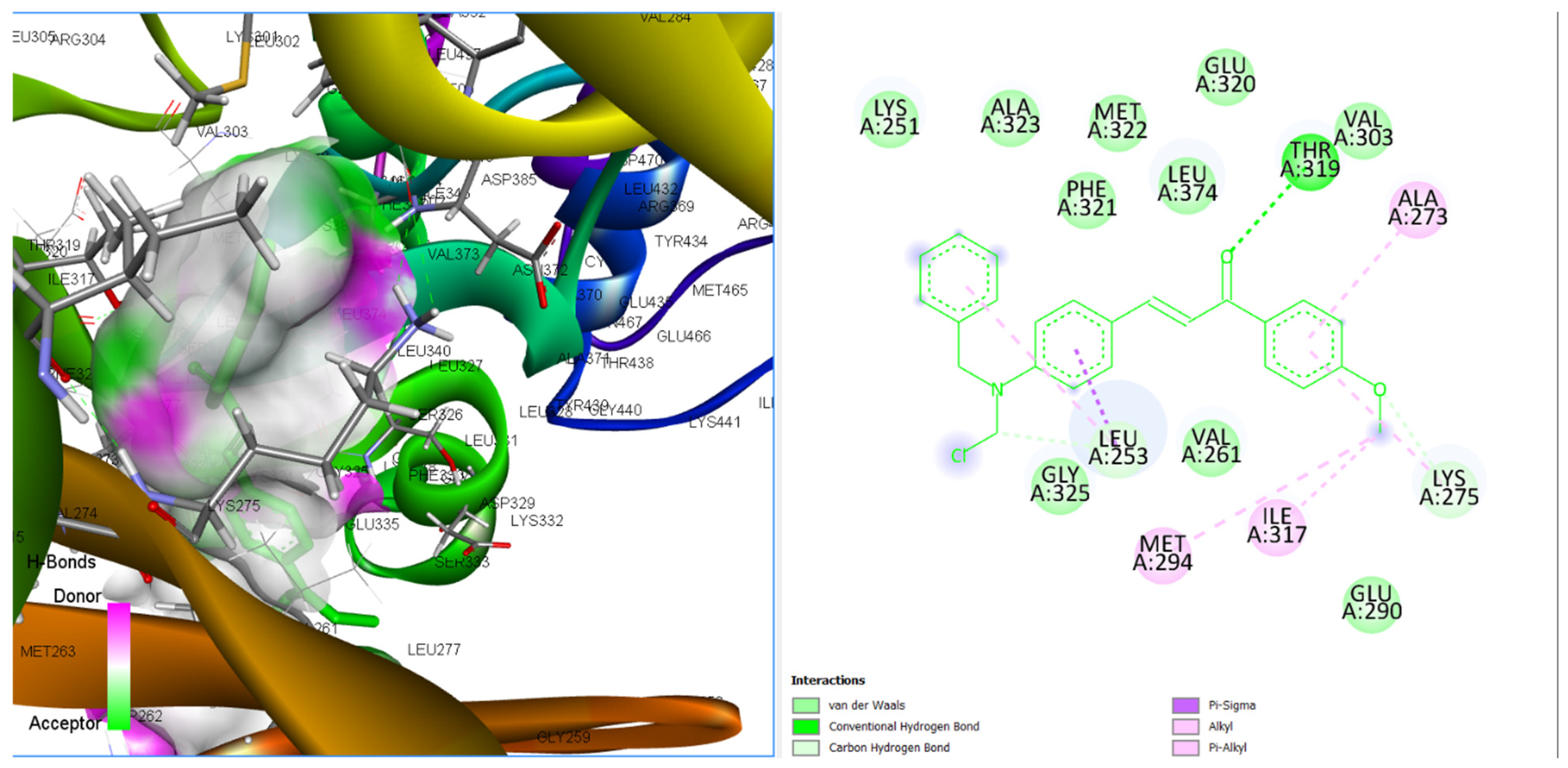
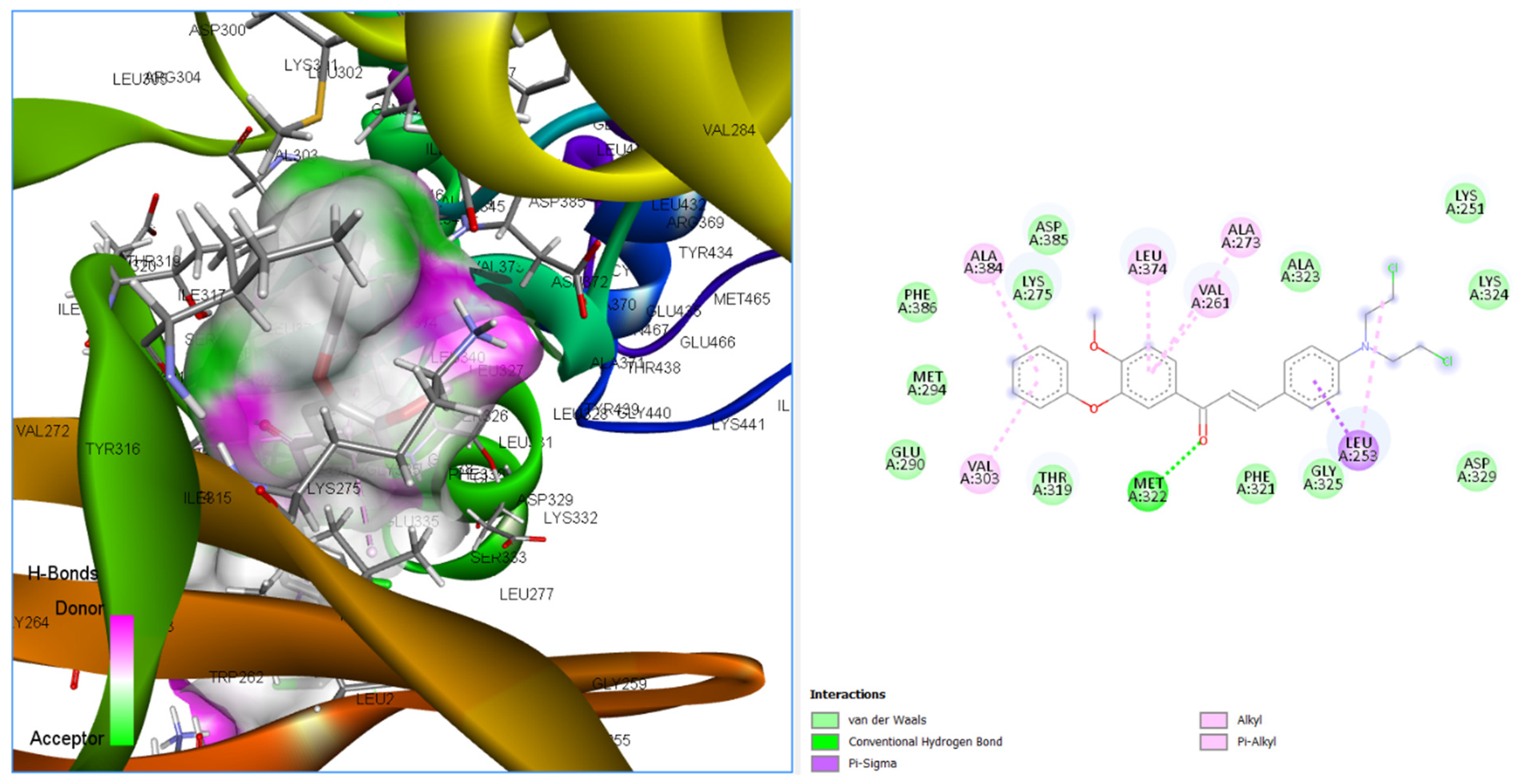
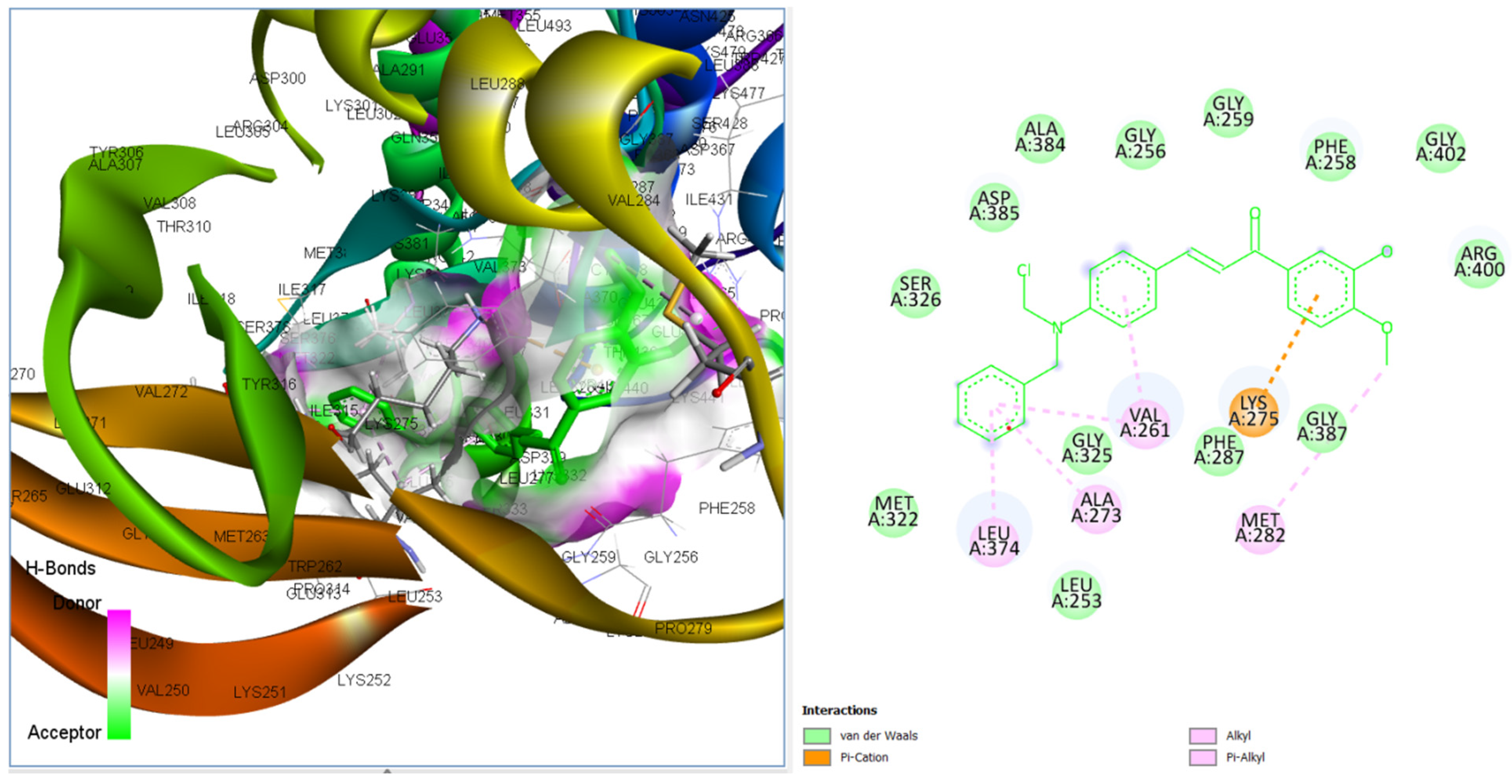
3.3.4. Binding Poses and Binding Interaction Analysis of Designed Compounds Against Lyn Tyrosine Kinase Enzyme
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment. JAMA 2019, 321, 288. [Google Scholar] [CrossRef] [PubMed]
- Almansour, N.M. Triple-Negative Breast Cancer: A Brief Review About Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, K.; Baumann, C.; Szabadkai, I.; Őrfi, L.; Kéri, G.; Ullrich, A.; Torka, R. Combined inhibition of AXL, Lyn and p130Cas kinases block migration of triple negative breast cancer cells. Cancer Biol. Ther. 2014, 15, 1571–1582. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.L.; Bocanegra, M.; Kwon, M.J.; Shin, Y.K.; Nam, S.J.; Yang, J.H.; Kao, J.; Godwin, A.K.; Pollack, J.R. LYN Is a Mediator of Epithelial-Mesenchymal Transition and a Target of Dasatinib in Breast Cancer. Cancer Res. 2010, 70, 2296–2306. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors: A 2024 update. Pharmacol. Res. 2024, 200, 107059. [Google Scholar] [CrossRef] [PubMed]
- Elkhalifa, D.; Alali, F.; Al Moustafa, A.-E.; Khalil, A. Targeting triple negative breast cancer heterogeneity with chalcones: A molecular insight. J. Drug Target. 2019, 27, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Singh, R.; Sharma, D.; Parvaiz Hassan, Q.; Gopu, B.; Momo, H.; Anal, J. Design, Synthesis and Biological Evaluation of Chalcone Acetamide Derivatives against Triple Negative Breast Cancer. Bioorganic Med. Chem. Lett. 2023, 107, 129795. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kontoyianni, M. Docking and Virtual Screening in Drug Discovery. In Proteomics for Drug Discovery: Methods and Protocols; Springer: New York, NY, USA, 2017; pp. 255–266. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed]
- Weerawarna, P.M.; Richardson, T.I. Lyn Kinase Structure, Regulation, and Involvement in Neurodegenerative Diseases: A Mini Review. Kinases Phosphatases 2023, 1, 23–38. [Google Scholar] [CrossRef]
| Compounds | ||
|---|---|---|
 |  |  |
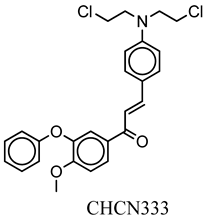 | 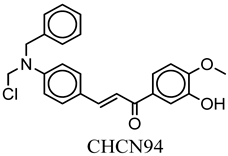 | |
| Compound Name | CHCN1 | CHCN19 | CHCN48 | CHCN333 | CHCN94 |
|---|---|---|---|---|---|
| Formula | C26H24Cl3NO2 | C20H21Cl2NO3 | C24H22ClNO2 | C26H25Cl2NO3 | C24H22ClNO3 |
| Molecular weight | 488.83 g/mol | 394.29 g/mol | 391.89 g/mol | 470.39 g/mol | 407.89 g/mol |
| Num. heavy atoms | 32 | 26 | 28 | 32 | 29 |
| Num. arom. heavy atoms | 18 | 12 | 18 | 18 | 18 |
| Fraction Csp3 | 0.19 | 0.255 | 0.12 | 0.19 | 0.12 |
| Num. rotatable bonds | 10 | 9 | 8 | 11 | 8 |
| Num. H-bond acceptors | 2 | 3 | 2 | 3 | 3 |
| Num. H-bond donors | 0 | 1 | 0 | 0 | 1 |
| Molar Refractivity | 136.60 | 108.18 | 116.23 | 132.67 | 118.25 |
| TPSA | 29.54 Å2 | 49.77 Å2 | 29.54 Å2 | 38.77 Å2 | 49.77 Å2 |
| Log Po/w (MLOGP) | 5.28 | 3.22 | 4.21 | 4.50 | 3.61 |
| Inference | Yes | Yes | Yes | Yes | Yes |
| Lipinski’s Violation | 1 | 0 | 0 | 0 | 0 |
| Veber Violation | 0 | 0 | 0 | 1 | 0 |
| Ghose violations | 3 | 0 | 0 | 2 | 0 |
| Egan Violation | 1 | 0 | 0 | 1 | 0 |
| Muegge Violation | 1 | 1 | 1 | 1 | 1 |
| Bioavailability Score | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| Synthetic accessibility | 3.38 | 2.94 | 2.90 | 3.45 | 3.01 |
| Compound Name | CHCN1 | CHCN19 | CHCN48 | CHCN333 | CHCN94 |
|---|---|---|---|---|---|
| Silicos-IT LogSw | −10.23 | −6.62 | −8.29 | −6.54 | −7.70 |
| Silicos-class | Insoluble | Poorly Soluble | Poorly soluble | Poorly soluble | Poorly soluble |
| Log Kp (cm/s) | −4.16 cm/s | −5.04 cm/s | −4.45 cm/s | −4.56 cm/s | −4.81 cm/s |
| GI Absorption | Low | High | High | High | High |
| BBB Permeant | No | Yes | Yes | No | Yes |
| Pgp substrate | Yes | No | Yes | Yes | Yes |
| CYP1A2 inhibitor | Yes | Yes | Yes | Yes | Yes |
| CYP2C19 inhibitor | No | Yes | Yes | No | Yes |
| CYP2C9 inhibitor | No | Yes | Yes | Yes | Yes |
| CYP2D6 inhibitor | No | Yes | Yes | Yes | Yes |
| CYP3A4 inhibitor | Yes | Yes | Yes | Yes | Yes |
| Properties | CHCN1 | CHCN19 | CHCN48 | CHCN333 | CHCN94 |
|---|---|---|---|---|---|
| Oral acute toxicity | V | V | IV | IV | IV |
| Carcinogenicity | + | − | − | − | − |
| Hepatotoxicity | − | + | + | + | − |
| Androgen receptor binding | − | − | − | − | − |
| Thyroid receptor Binding | − | − | − | − | − |
| Estrogen receptor Binding | − | + | + | + | + |
| Aromatase binding | − | + | + | + | + |
| Compounds | Hydrogen Bond Interaction | Hydrophobic Interaction |
|---|---|---|
| CHCN1 | THR319 MET322 GLU320 LEU253 | ALA384 LYS251 LEU253 VAL261 ALA273 LEU374 LEU253 |
| CHCN19 | THR319 LYS275 LEU253 LEU253 | LEU253 MET294 ILE317 ALA273 LYS275 LEU253 |
| CHCN48 | THR319 LYS275 LEU253 | LEU253 MET294 ILE317 ALA273 LYS275 LEU253 |
| CHCN333 | MET322 | LEU253 LEU253 VAL261 ALA273 LEU374 VAL303 ALA384 |
| CHCN94 | MET282 VAL261 VAL261 ALA273 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onaji, E.; Jimoh, Y.; Hamza, A.N.; Abdullahi, M. Novel Chalcone Derivatives as Potent Lyn Tyrosine Kinase Inhibitors: A Promising In-Silico Approach for Targeted Therapy in Triple-Negative Breast Cancer. Chem. Proc. 2025, 18, 8. https://doi.org/10.3390/ecsoc-29-26887
Onaji E, Jimoh Y, Hamza AN, Abdullahi M. Novel Chalcone Derivatives as Potent Lyn Tyrosine Kinase Inhibitors: A Promising In-Silico Approach for Targeted Therapy in Triple-Negative Breast Cancer. Chemistry Proceedings. 2025; 18(1):8. https://doi.org/10.3390/ecsoc-29-26887
Chicago/Turabian StyleOnaji, Enayi, Yusuf Jimoh, Asmau Nasir Hamza, and Maryam Abdullahi. 2025. "Novel Chalcone Derivatives as Potent Lyn Tyrosine Kinase Inhibitors: A Promising In-Silico Approach for Targeted Therapy in Triple-Negative Breast Cancer" Chemistry Proceedings 18, no. 1: 8. https://doi.org/10.3390/ecsoc-29-26887
APA StyleOnaji, E., Jimoh, Y., Hamza, A. N., & Abdullahi, M. (2025). Novel Chalcone Derivatives as Potent Lyn Tyrosine Kinase Inhibitors: A Promising In-Silico Approach for Targeted Therapy in Triple-Negative Breast Cancer. Chemistry Proceedings, 18(1), 8. https://doi.org/10.3390/ecsoc-29-26887






