Molecular Docking/ADME-TOX-Based Analysis for New Anti-Colorectal Cancer Through Peroxiredoxin 1 Inhibition †
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Molecular Docking
3.2. Evaluation ADME-TOX
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adebayo, A.S.; Agbaje, K.; Adesina, S.K.; Olajubutu, O. Colorectal Cancer: Disease Process, Current Treatment Options, and Future Perspectives. Pharmaceutics 2023, 15, 2620. [Google Scholar] [CrossRef] [PubMed]
- Marcellinaro, R.; Spoletini, D.; Grieco, M.; Avella, P.; Cappuccio, M.; Troiano, R.; Lisi, G.; Garbarino, G.M.; Carlini, M. Colorectal Cancer: Current Updates and Future Perspectives. J. Clin. Med. 2024, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Bardelčíková, A.; Šoltys, J.; Mojžiš, J. Oxidative Stress, Inflammation and Colorectal Cancer: An Overview. Antioxidants 2023, 12, 901. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.R.; Rosa, I.; Claro, I. Early-onset colorectal cancer: A review of current knowledge. World J. Gastroenterol. 2023, 29, 1289–1303. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Eglinton, T.W.; Frizelle, F.A.; Hampton, M.B. Peroxiredoxins in Colorectal Cancer: Predictive Biomarkers of Radiation Response and Therapeutic Targets to Increase Radiation Sensitivity? Antioxidants 2018, 7, 136. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Tan, W.; Yang, Q.; Zou, Z.; Zhou, R.; Huang, Y.; Qiu, Z.; Zheng, K.; Huang, Z. PRRX1 promotes colorectal cancer stemness and chemoresistance via the JAK2/STAT3 axis by targeting IL-6. J. Gastrointest. Oncol. 2022, 13, 2989–3008. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, H.; Ding, C.; Jiang, D.; Zhao, Z.; Li, Y.; Ding, X.; Gao, J.; Zhou, H.; Luo, C.; et al. Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1. Signal Transduct. Target. Ther. 2023, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Thapa, P.; Jiang, H.; Ding, N.; Hao, Y.; Alshahrani, A.; Wei, Q. The Role of Peroxiredoxins in Cancer Development. Biology 2023, 12, 666. [Google Scholar] [CrossRef]
- Rani, P.; Chahal, S.; Kumar, R.; Mayank, K.P.; Negi, A.; Singh, R.; Kumar, S.; Kataria, R.; Joshi, G.; Sindhu, J. Electro-organic synthesis of C-5 sulfenylated amino uracils: Optimization and exploring topoisomerase-I based anti-cancer profile. Bioorganic Chem. 2023, 138, 106660. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.W.; Huang, W.J.; Liu, Y.H.; Liu, Q.G.; Song, J.; Hu, T.; Chen, P.; Zhang, S.Y. Design, synthesis and biological evaluation of 1,2,3-triazole benzothiazole derivatives as tubulin polymerization inhibitors with potent anti-esophageal cancer activities. Eur. J. Med. Chem. 2024, 265, 116118. [Google Scholar] [CrossRef] [PubMed]
- HyperChem(TM) Professional 7.51; Hypercube, Inc.: Gainesville, FL, USA, 2001.
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Saghiri, K.; Daoud, I.; Melkemi, N.; Mesli, F. Molecular docking/dynamics simulations, MEP analysis, and pharmacokinetics prediction of some withangulatin A derivatives as allosteric glutaminase C inhibitors in breast cancer. Chem. Data Collect. 2023, 46, 101044. [Google Scholar] [CrossRef]
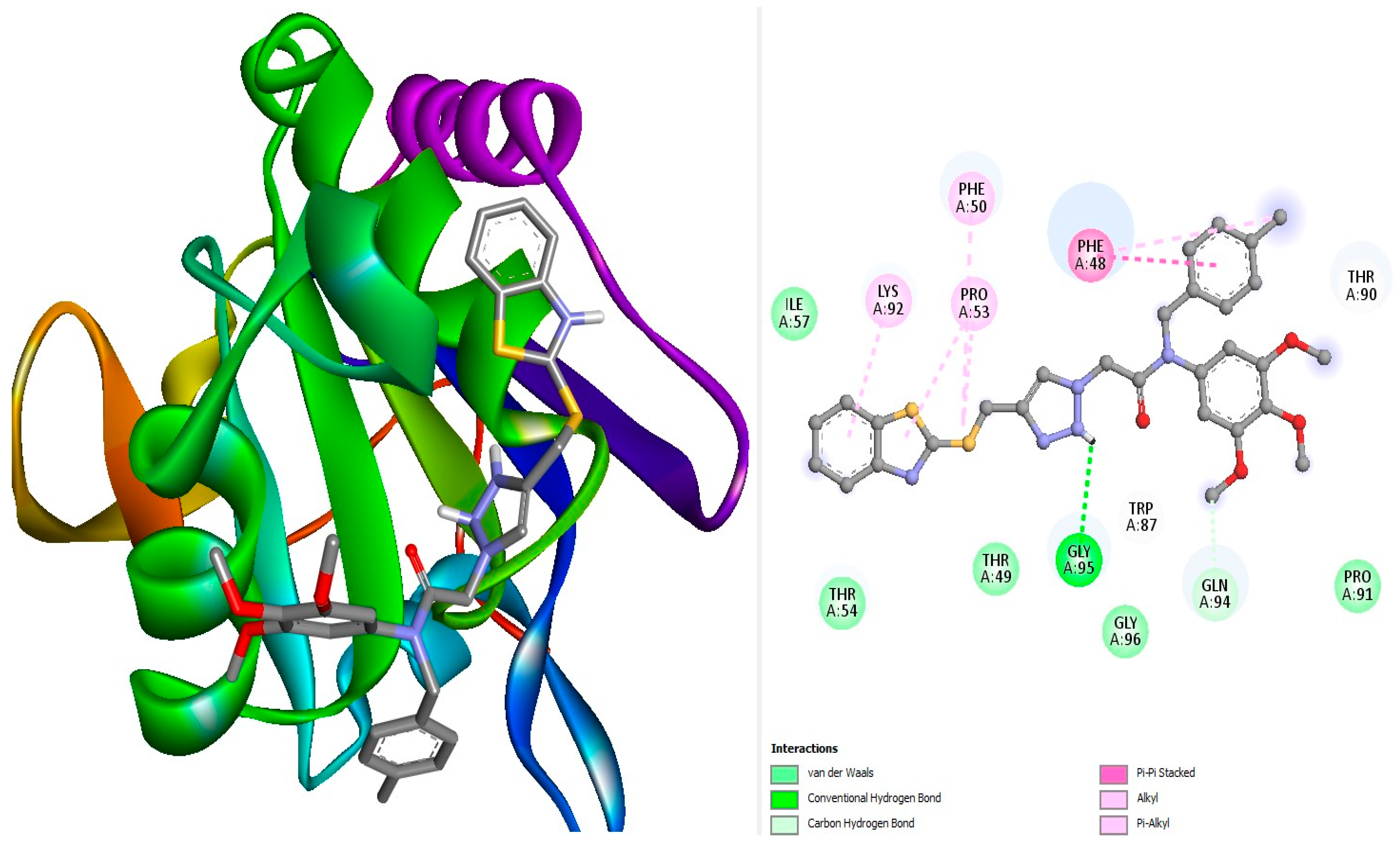
| Complex | Binding Affinity (kcal/Mol) | Bonds Between the Compounds, Atoms, and Active Site Residues (Chain A) | ||||
|---|---|---|---|---|---|---|
| Interaction Type | Receptor Residues | Receptor Atoms | Compound Atoms | Distance (A°) | ||
| Ref1 | −6.8 | Pi-Pi stacked | Phe50 | 6-ring | 6-ring | 4.4 |
| L18 | −7.4 | H-Bond H-Bond | Gly95 Gln94 | O O | NH H5 | 2.28 2.97 |
| L17 | −7.3 | H-Bond H-Bond | Gly95 Thr49 | O O | NH NH | 2.28 2.59 |
| L25 | −7.3 | H-Bond H-Bond | Gly95 Thr49 | O O | NH NH | 2.22 2.65 |
| L19 | −7.2 | H-Bond H-Bond | Gly95 Gly94 | O O | NH C | 2.19 3.5 |
| L20 | −7.2 | H-Bond | Gly95 | O | NH | 2.1 |
| L26 | −7.2 | H-Bond | Gly95 | O | NH | 2.07 |
| L22 | −7.0 | H-Bond | Gly95 | O | NH | 2.13 |
| L23 | −7.0 | H-Bond | Gly95 | O | NH | 2.24 |
| L24 | −7.0 | H-Bond | Gly95 | O | NH | 2.18 |
| Category | Model Name | L25 | L20 | L26 |
|---|---|---|---|---|
| Absorption | Water solubility | −5.57 | −4.91 | −5.57 |
| Caco-2 permeability | 0.75 | 0.98 | 0.75 | |
| HIA (%absorbed) | 83.3 | 90.3 | 83.6 | |
| P-gp substrate | Yes | Yes | Yes | |
| P-gp I inhibitor | Yes | Yes | Yes | |
| Distribution | VDss | −0.25 | −0.34 | −0.28 |
| BBB permeability | −2.01 | −1.39 | −1.99 | |
| Metabolism | CYP2D6 substrate | No | No | No |
| CYP3A4 substrate | Yes | Yes | Yes | |
| CYP1A2 inhibitor | No | No | No | |
| CYP2C19 inhibitor | Yes | Yes | Yes | |
| CYP2C9 inhibitor | Yes | Yes | Yes | |
| CYP2D6 inhibitor | No | No | No | |
| CYP3A4 inhibitor | Yes | Yes | Yes | |
| Excretion | Total clearance | 0.76 | 0.73 | 0.78 |
| T1/2 (h) | 1.14 | 0.72 | 1.06 | |
| Toxicity | AMES toxicity | No | No | No |
| Maximum tolerated dose (log mg/kg/day) | 0.69 | 0.68 | 0.69 | |
| HERG I inhibitor | No | No | No | |
| HERG II inhibitor | Yes | Yes | Yes | |
| Oral rat acute toxicity (LD50) (mol/kg) | 2.41 | 1.75 | 2.41 | |
| Hepatotoxicity | No | No | No | |
| Skin sensitization | No | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bensahbane, I.; Melkemi, N.; Daoud, I.; Asli, F. Molecular Docking/ADME-TOX-Based Analysis for New Anti-Colorectal Cancer Through Peroxiredoxin 1 Inhibition. Chem. Proc. 2024, 16, 56. https://doi.org/10.3390/ecsoc-28-20215
Bensahbane I, Melkemi N, Daoud I, Asli F. Molecular Docking/ADME-TOX-Based Analysis for New Anti-Colorectal Cancer Through Peroxiredoxin 1 Inhibition. Chemistry Proceedings. 2024; 16(1):56. https://doi.org/10.3390/ecsoc-28-20215
Chicago/Turabian StyleBensahbane, Imane, Nadjib Melkemi, Ismail Daoud, and Faiza Asli. 2024. "Molecular Docking/ADME-TOX-Based Analysis for New Anti-Colorectal Cancer Through Peroxiredoxin 1 Inhibition" Chemistry Proceedings 16, no. 1: 56. https://doi.org/10.3390/ecsoc-28-20215
APA StyleBensahbane, I., Melkemi, N., Daoud, I., & Asli, F. (2024). Molecular Docking/ADME-TOX-Based Analysis for New Anti-Colorectal Cancer Through Peroxiredoxin 1 Inhibition. Chemistry Proceedings, 16(1), 56. https://doi.org/10.3390/ecsoc-28-20215






