Multi-Target In Silico Evaluation of New 2-Pyrazolines as Antimicrobial Agents †
Abstract
1. Introduction
2. Methods
3. Results and Discussion
3.1. Drug-Likeness and Bioavailability
3.2. Synthetic Accessibility
3.3. LogP and Water Solubility
3.4. Binding Affinity and Ligand–Receptor Interaction
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boudou, F.; Sehmi, A.; Belakredar, A.; Zaoui, O. Synthesis, characterization, antimicrobial activity, and in silico assessment of a novel pyrazoline carboxamide heterocyclic compound. Bangladesh J. Pharmacol. 2023, 18, 152–161. [Google Scholar] [CrossRef]
- El-Naggar, M.; Rashdan HR, M.; Abdelmonsef, A.H. Cyclization of Chalcone Derivatives: Design, Synthesis, In Silico Docking Study, and Biological Evaluation of New Quinazolin-2,4-diones Incorporating Five-, Six-, and Seven-Membered Ring Moieties as Potent Antibacterial Inhibitors. ACS Omega 2023, 8, 27216–27230. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Rasoo, M.H.; Nisar, M.A.; Aslam, M.A.; Qamar, M.U. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Bajorath, J. Designing highly potent compounds using a chemical language model. Sci. Rep. 2023, 13, 7412–7423. [Google Scholar] [CrossRef] [PubMed]
- Raauf, A.M.R.; Omar, T.N.A.; Mahdi, M.F.; Fadhil, H.R. Synthesis, molecular docking and anti-inflammatory evaluation of new trisubstituted pyrazoline derivatives bearing benzenesulfonamide moiety. Nat. Prod. Res. 2024, 38, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Ebenezer, O.; Shapi, M.; Tuszynski, J.A. A review of the recent development in the synthesis and biological evaluations of pyrazole derivatives. Biomedicines 2022, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.H.; Abdula, A.M.; Tomi, I.H.; Al-Daraji, A.H.; Baqi, Y. Synthesis, Antimicrobial Evaluation and Docking Study of Novel 3,5-Disubstituted-2-Isoxazoline and 1,3,5-Trisubstituted-2-Pyrazoline Derivatives. Med. Chem. 2021, 17, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, C.; Zhang, L.; Sun, B.; Cui, Y.; Sang, F. Isolation and biological activity of natural chalcones based on antibacterial mechanism classification. Bioorganic. Med. Chem. 2023, 93, 117454. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.J. Computational Development of Inhibitors of Plasmid-Borne Bacterial Dihydrofolate Reductase. Antibiotics 2022, 11, 779. [Google Scholar] [CrossRef]
- Cui, T.M.; Altaf, M.; Aldarhami, A.; Bazaid, A.S.; Saeedi, N.H.; Alkayyal, A.A.; Alshabrmi, F.M.; Ali, F.; Aladhadh, M.; Khan, M.Y.; et al. Dihydropyrimidone Derivatives as Thymidine Phosphorylase Inhibitors: Inhibition Kinetics, Cytotoxicity, and Molecular Docking. Molecules 2023, 28, 3634. [Google Scholar] [CrossRef] [PubMed]
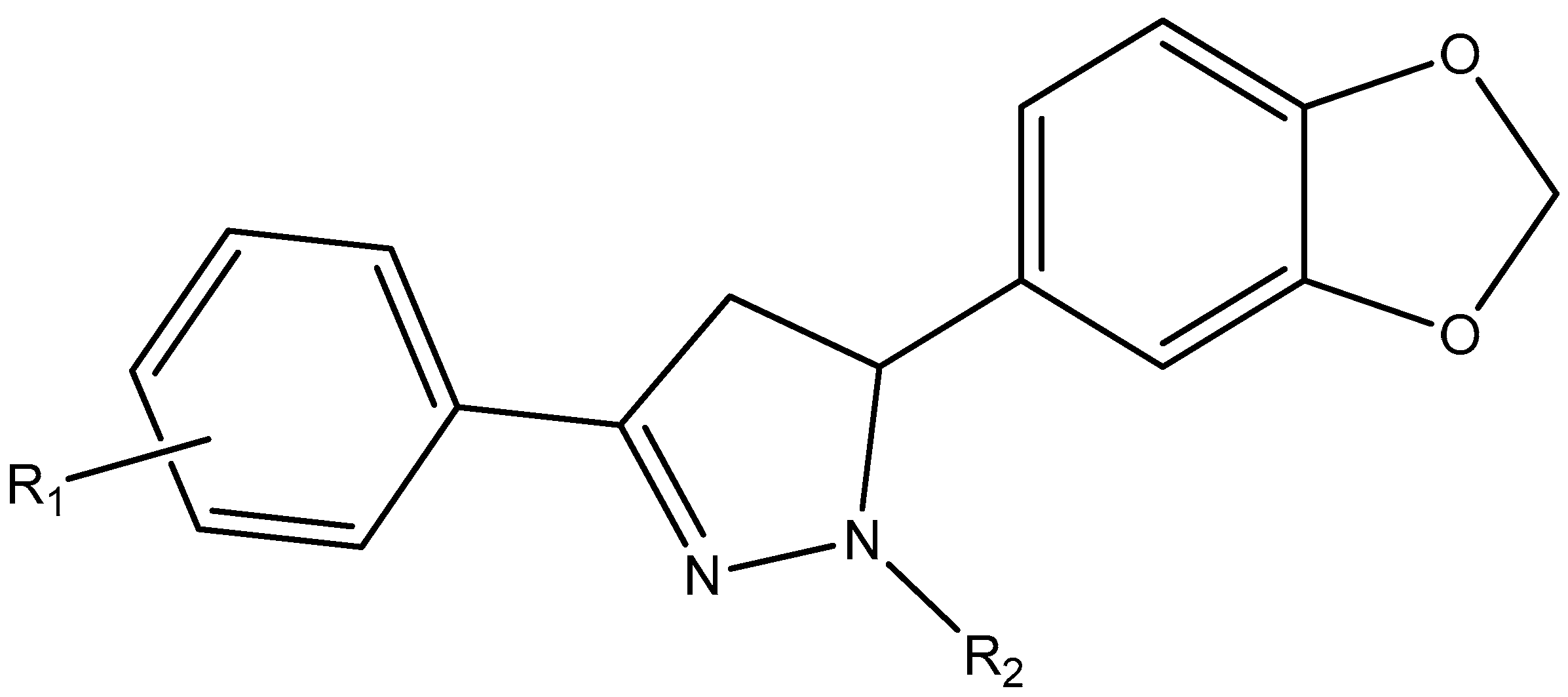
| Lig | Formula | MW | H-Acceptors | H-Bond Donors | Water Solubility | XLOGP3 | Bioavailability Score | Synthetic Accessibility |
|---|---|---|---|---|---|---|---|---|
| 2 | C18H17N3O3 | 323.35 | 4 | 1 | Soluble | 2.31 | 0.55 | 3.57 |
| 3 | C19H18N2O3 | 322.36 | 4 | 0 | Soluble | 2.88 | 0.55 | 3.54 |
| 5 | C23H20N2O2 | 356.42 | 3 | 0 | Moderately soluble | 5.2 | 0.55 | 3.7 |
| 7 | C19H19N3O3 | 337.37 | 4 | 1 | Soluble | 2.67 | 0.55 | 3.69 |
| 8 | C20H20N2O3 | 336.38 | 4 | 0 | Soluble | 3.24 | 0.55 | 3.66 |
| 9 | C20H20N2O4 | 352.38 | 5 | 0 | Moderately soluble | 3.65 | 0.55 | 3.8 |
| 10 | C24H22N2O2 | 370.44 | 3 | 0 | Poorly soluble | 5.57 | 0.55 | 3.82 |
| 12 | C17H14ClN3O3 | 343.76 | 4 | 1 | Soluble | 2.57 | 0.55 | 3.46 |
| 13 | C18H15ClN2O3 | 342.78 | 4 | 0 | Soluble | 3.14 | 0.55 | 3.43 |
| 15 | C22H17ClN2O2 | 376.84 | 3 | 0 | Moderately soluble | 5.46 | 0.55 | 3.6 |
| 17 | C17H13Cl2N3O3 | 378.21 | 4 | 1 | Moderately soluble | 3.2 | 0.55 | 3.48 |
| 18 | C18H14Cl2N2O3 | 377.22 | 4 | 0 | Moderately soluble | 3.77 | 0.55 | 3.45 |
| 19 | C18H14Cl2N2O4 | 393.22 | 5 | 0 | Moderately soluble | 4.17 | 0.55 | 3.58 |
| 20 | C22H16Cl2N2O2 | 411.28 | 3 | 0 | Poorly soluble | 6.09 | 0.55 | 3.61 |
| 25 | C22H18N2O3 | 358.39 | 4 | 1 | Moderately soluble | 4.48 | 0.55 | 3.64 |
| 27 | C18H16N2O5 | 340.33 | 6 | 2 | Soluble | 1.8 | 0.55 | 3.53 |
| 29 | C18H16N2O6 | 356.33 | 7 | 2 | Soluble | 2.21 | 0.55 | 3.67 |
| 30 | C22H18N2O4 | 374.39 | 5 | 2 | Moderately soluble | 4.13 | 0.55 | 3.7 |
| 35 | C23H20N2O3 | 372.42 | 4 | 0 | Moderately soluble | 4.81 | 0.55 | 3.75 |
| 40 | C24H22N2O4 | 402.44 | 5 | 0 | Moderately soluble | 4.78 | 0.55 | 3.93 |
| LIGANDS | SUBSTITUENTS | REPORT | PROTOX CLASS | BINDING ENERGY | |||
|---|---|---|---|---|---|---|---|
| R1 | R2 | 1MOQ | 4EAD | 7MQP | |||
| NATIVE LIGAND | −7.5 | −6.8 | −9.6 | ||||
| AMOXYCILLIN | −7.9 | −8.6 | −6.8 | ||||
| 2 | 4-CH3 | ONH2 | REPORTED | 4 | −8.9 | −8.7 | −9.2 |
| 3 | 4-CH3 | (CO)CH3 | REPORTED | 4 | −8.9 | −8.7 | −9.2 |
| 5 | 4-CH3 | C6H5 | NEW | 4 | −7.9 | −9.5 | −9.9 |
| 7 | 3,5-CH3 | ONH2 | NEW | 4 | −9.1 | −8.9 | −9.3 |
| 8 | 3,5-CH3 | (CO)CH3 | NEW | 4 | −9.2 | −9.0 | −9.8 |
| 9 | 3,5-CH3 | (CO)OCH3 | NEW | 4 | −8.7 | −8.9 | −9.1 |
| 10 | 3,5-CH3 | C6H5 | NEW | 4 | −7.9 | −9.6 | −9.2 |
| 12 | 4-Cl | ONH2 | NEW | 4 | −8.8 | −8.2 | −9.2 |
| 13 | 4-Cl | (CO)CH3 | NEW | 4 | −8.8 | −8.6 | −9.1 |
| 15 | 4-Cl | C6H5 | NEW | 4 | −7.9 | −9.3 | −9.8 |
| 17 | 3,5-Cl | ONH2 | NEW | 4 | −8.9 | −8.5 | −9.2 |
| 18 | 3,5-Cl | (CO)CH3 | NEW | 4 | −9.0 | −8.7 | −9.5 |
| 19 | 3,5-Cl | (CO)OCH3 | NEW | 4 | −8.7 | −8.7 | −9.5 |
| 20 | 3,5-Cl | C6H5 | NEW | 4 | −7.8 | −9.4 | −10.1 |
| 25 | 4-OH | C6H5 | NEW | 4 | −7.6 | −9.1 | −9.2 |
| 27 | 3,5-OH | ONH2 | NEW | 4 | −8.6 | −8.7 | −9.5 |
| 29 | 3,5-OH | (CO)OCH3 | NEW | 4 | −8.2 | −8.7 | −9.3 |
| 30 | 3,5-OH | C6H5 | NEW | 4 | −8.2 | −9.2 | −10.2 |
| 35 | 4-(CO)OCH3 | C6H5 | NEW | 4 | −7.5 | −9.1 | −8.6 |
| 40 | 3,5-(CO)OCH3 | C6H5 | NEW | 4 | −7.8 | −8.9 | −8.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salami, Z.; Hamza, A.; Idris, A.; Jimoh, Y. Multi-Target In Silico Evaluation of New 2-Pyrazolines as Antimicrobial Agents. Chem. Proc. 2024, 16, 110. https://doi.org/10.3390/ecsoc-28-20226
Salami Z, Hamza A, Idris A, Jimoh Y. Multi-Target In Silico Evaluation of New 2-Pyrazolines as Antimicrobial Agents. Chemistry Proceedings. 2024; 16(1):110. https://doi.org/10.3390/ecsoc-28-20226
Chicago/Turabian StyleSalami, Zukhruf, Asmau Hamza, Abdullahi Idris, and Yusuf Jimoh. 2024. "Multi-Target In Silico Evaluation of New 2-Pyrazolines as Antimicrobial Agents" Chemistry Proceedings 16, no. 1: 110. https://doi.org/10.3390/ecsoc-28-20226
APA StyleSalami, Z., Hamza, A., Idris, A., & Jimoh, Y. (2024). Multi-Target In Silico Evaluation of New 2-Pyrazolines as Antimicrobial Agents. Chemistry Proceedings, 16(1), 110. https://doi.org/10.3390/ecsoc-28-20226






