Carvacrol: A PLpro Inhibitor of SARS-CoV-2 Is a Natural Weapon for COVID-19 †
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Ligand Preparation
2.3. Protein Preparation and Receptor Grid Generation
2.4. Molecular Docking Study
2.5. In Silico Prediction of Absorption, Distribution, Metabolism, and Excretion (ADME)
3. Result and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Arden, K.E.; Nissen, M.D.; Sloots, T.P.; Mackay, I.M. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J. Med. Virol. 2005, 75, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.-M.; Chan, K.-H.; Tsoi, H.-W.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.-K.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef] [PubMed]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent Insights into Emerging Coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Haleem, A.; Javaid, M.; Vaishya, R. Effects of COVID-19 pandemic in daily life. Curr. Med. Res. Pract. 2020, 10, 78–79. [Google Scholar] [CrossRef]
- Hoofman, J.; Secord, E. The Effect of COVID-19 on Education. Pediatr. Clin. N. Am. 2021, 68, 1071–1079. [Google Scholar] [CrossRef]
- Mustafa, N. Impact of the 2019–20 Coronavirus Pandemic on Education. Available online: https://www.researchgate.net/publication/340849956_Impact_of_the_2019-20_coronavirus_pandemic_on_education?channel=doi&linkId=5ea0ae4c299bf143893ff447&showFulltext=true (accessed on 20 May 2021).
- United Nations Educational, Scientific and Cultural Organization (UNESCO). Adverse Consequences of School Closures. 10 March 2020. Available online: https://en.unesco.org/covid19/educationresponse/consequences (accessed on 20 May 2021).
- Hussain, W.; Haleem, K.S.; Khan, I.; Tauseef, I.; Qayyum, S.; Ahmed, B.; Riaz, M.N. Medicinal plants: A repository of antiviral metabolites. Future Virol. 2017, 12, 299–308. [Google Scholar] [CrossRef]
- Denaro, M.; Smeriglio, A.; Barreca, D.; De Francesco, C.; Occhiuto, C.; Milano, G.; Trombetta, D. Antiviral activity of plants and their isolated bioactive compounds: An update. Phytother. Res. 2019, 34, 742–768. [Google Scholar] [CrossRef]
- Lin, L.-T.; Hsu, W.-C.; Lin, C.-C. Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med. 2014, 4, 24–35. [Google Scholar] [CrossRef]
- Ibrahim, M.A.A.; Abdelrahman, A.H.M.; Hussien, T.A.; Badr, E.A.A.; Mohamed, T.A.; El-Seedi, H.R.; Pare, P.W.; Efferth, T.; Hegazy, M.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020, 126, 104046. [Google Scholar] [CrossRef]
- Ibrahim, M.A.A.; Abdeljawaad, K.A.A.; Abdelrahman, A.H.M.; Hegazy, M.E.F. Natural-like products as potential SARS-CoV-2 Mpro inhibitors: In-silico drug discovery. J. Biomol. Struct. Dyn. 2021, 39, 5722–5734. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Debnath, B.; Debnath, P.; Debnath, S.; Zaki, M.E.A.; Masand, V.H. Identification of potential edible mushroom as SARS-CoV-2 main protease inhibitor using rational drug design approach. Sci. Rep. 2022, 12, 1503. [Google Scholar] [CrossRef]
- Sen, D.; Debnath, P.; Debnath, B.; Bhaumik, S.; Debnath, S. Identification of potential inhibitors of SARS-CoV-2 main protease and spike receptor from 10 important spices through structure-based virtual screening and molecular dynamic study. J. Biomol. Struct. Dyn. 2020, 40, 941–962. [Google Scholar] [CrossRef] [PubMed]
- Khaerunnisa, S.; Kurniawan, H.; Awaluddin, R.; Suhartati, S.; Soetjipto, S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints 2020, 2020, 2020030226. [Google Scholar] [CrossRef]
- Shamim Molla, M.; Azad, A.K.; Al Hasib, M.A.A.; Hossain, M.M.; Ahammed, M.S.; Rana, S.; Islam, M.T. A review on antiviral effects of Nigella Sativa L. Pharmacol. Online 2019, 2, 47–53. [Google Scholar]
- Basurra, R.S.; Wang, S.M.; Alhoot, M.A. Nigella sativa (Black Seed) as a Natural Remedy against Viruses. J. Pure Appl. Microbiol. 2021, 15, 29–41. [Google Scholar] [CrossRef]
- Ahmad, S.; Abbasi, H.W.; Shahid, S.; Gul, S.; Abbasi, S.W. Molecular docking, simulation and MM-PBSA studies of Nigella sativa compounds: A computational quest to identify potential natural antiviral for COVID-19 treatment. J. Biomol. Struct. Dyn. 2021, 39, 4225–4233. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.L.; Hossain, M.S. Protective effect of black seed oil from Nigella sativa against murine cytomegalovirus infection. Int. J. Immunopharmacol. 2000, 22, 729–740. [Google Scholar] [CrossRef]
- Zaher, K.S.; Ahmed, W.M.; Zerizer, S.N. Observations on the biological effects of black cumin seed (Nigella sativa) and green tea (Camellia sinensis). Glob. Vet. 2008, 2, 198–204. [Google Scholar]
- Mukhtar, H.; Qureshi, A.S.; Anwar, F.; Mumtaz, M.W.; Marcu, M. Nigella sativa L. seed and seed oil: Potential sources of high-value components for development of functional foods and nutraceuticals/pharmaceuticals. J. Essent. Oil Res. 2019, 31, 171–183. [Google Scholar] [CrossRef]
- Islam, M.T.; Khan, M.R.; Mishra, S.K. An updated literature-based review: Phytochemistry, pharmacology and therapeutic promises of Nigella sativa L. Orient. Pharm. Exp. Med. 2019, 19, 115–129. [Google Scholar] [CrossRef]
- Ahmad, A.; Husain, A.; Mujeeb, M.; Alam Khan, S.; Najmi, A.K.; Siddique, N.A.; Damanhouri, Z.A.; Anwar, F. A review on therapeutic potential of Nigella sativa: A miracle herb. Asian Pac. J. Trop. Biomed. 2013, 3, 337–352. [Google Scholar] [CrossRef]
- Kokoska, L.; Havlik, J.; Valterova, I.; Sovova, H.; Sajfrtova, M.; Jankovska, I. Comparison of Chemical Composition and Antibacterial Activity of Nigella sativa Seed Essential Oils Obtained by Different Extraction Methods. J. Food Prot. 2008, 71, 2475–2480. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K. Cumin (Cuminum cyminum) and black cumin (Nigella sativa) seeds: Traditional uses, chemical constituents, and nutraceutical effects. Food Qual. Saf. 2018, 2, 1–16. [Google Scholar] [CrossRef]
- Debnath, B.; Debnath, P.; Ghosh, R.; Debnath, S. In Silico Identification of Potential Inhibitors of SARS-CoV-2 Papain-like Protease from Natural Sources: A Natural Weapon to Fight COVID-19. Coronaviruses 2020, 2, 16–27. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valiente, P.A.; Moreno, E. AMDock: A versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol. Direct 2020, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Schöning-Stierand, K.; Diedrich, K.; Fährrolfes, R.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Steinegger, R.; Rarey, M. ProteinsPlus: Interactive analysis of protein–ligand binding interfaces. Nucleic Acids Res. 2020, 48, W48–W53. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Yang, H.; Yang, M.; Ding, Y.; Liu, Y.; Lou, Z.; Zhou, Z.; Sun, L.; Mo, L.; Ye, S.; Pang, H.; et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 2003, 100, 13190–13195. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.B.; Jeong, H.J.; Kim, J.H.; Kim, Y.M.; Park, J.-Y.; Kim, D.; Naguyen, T.T.H.; Park, S.-J.; Chang, J.S.; Park, K.H.; et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010, 18, 7940–7947. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, R.; Yasuo, N.; Sekijima, M. Identification of key interactions between SARS-CoV-2 main protease and inhibitor drug candidates. Sci. Rep. 2020, 10, 12493. [Google Scholar] [CrossRef] [PubMed]
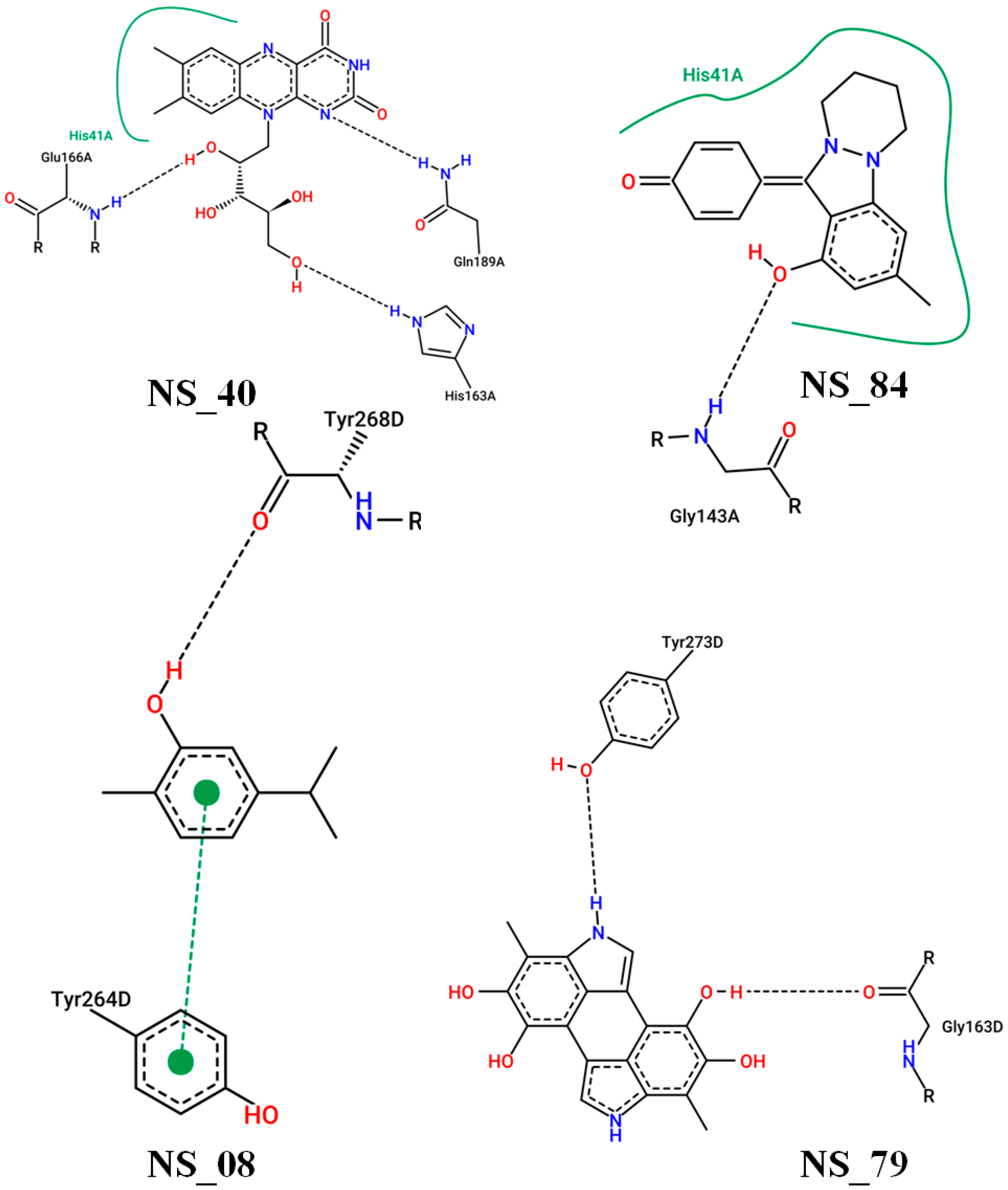
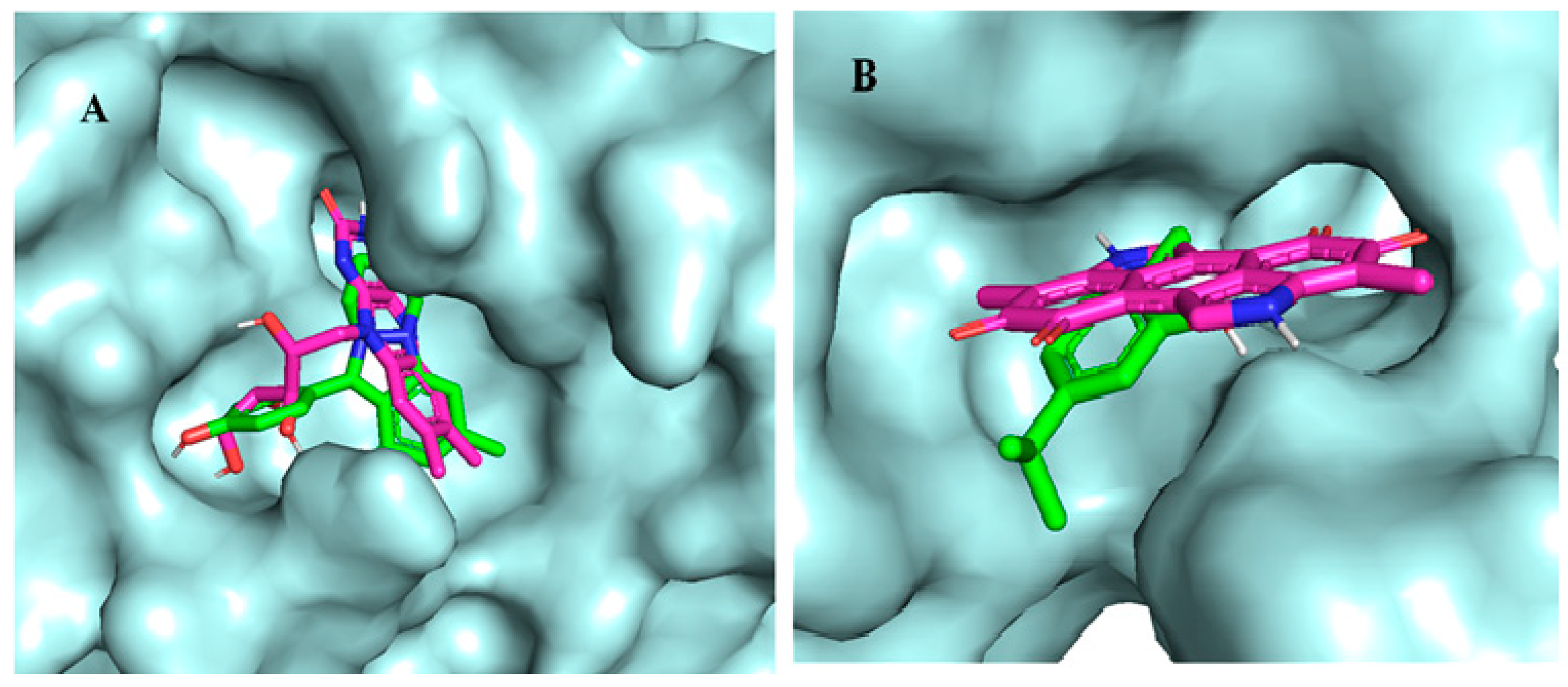
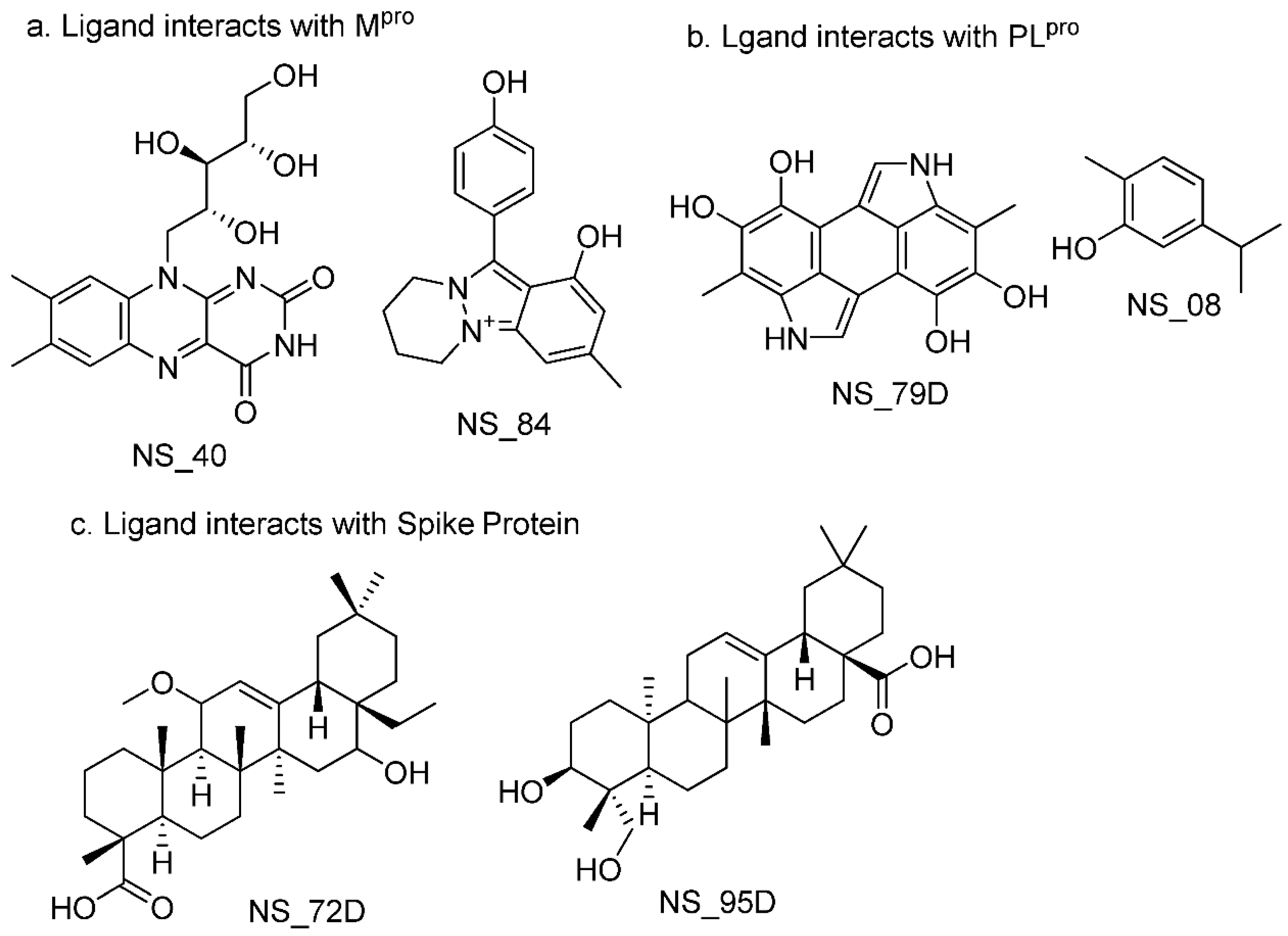
| Binding Affinity with Mpro | Binding Affinity with Spike | Binding Affinity with PLpro | |||
|---|---|---|---|---|---|
| Compound | Autodock Vina Binding Affinity (kcal/mol) | Compound | AutodockVina Binding Affinity (kcal/mol) | Compound | AutodockVina Binding Affinity (kcal/mol) |
| NS_21 | −6.7 | NS_21 | −7.4 | NS_79 | −6.4 |
| NS_40 | −7.5 | NS_40 | −6.1 | NS_70 | −5.7 |
| NS_66 | −6.5 | NS_72D | −7.7 | NS_93 | −5.7 |
| NS_84 | −7.9 | NS_95D | −7.5 | NS_102 | −5.5 |
| Carvacrol (NS_08) | −5.3 | Carvacrol (NS_08) | −5.1 | Carvacrol (NS_08) | −5.8 |
| Menthol | −5.0 | Menthol | −5.2 | Menthol | −5.6 |
| Parameters | NS_08 | NS_40 | NS_79 | NS_84 |
|---|---|---|---|---|
| MW | 150.22 | 376.36 | 318.28 | 295.36 |
| NHA | 11 | 27 | 24 | 22 |
| NAHA | 6 | 14 | 18 | 15 |
| NRB | 1 | 5 | 0 | 1 |
| NHBA | 1 | 8 | 4 | 2 |
| NHBD | 2 | 5 | 2 | 2 |
| MR | 40.01 | 96.99 | 93.74 | 88.27 |
| TPSA (Å2) | 20.23 | 161.56 | 99.86 | 49.27 |
| iLOGp | 2.24 | 0.91 | 0.97 | −0.57 |
| LogS (ESOL) | −3.31 | −1.59 | −4.60 | |
| MLOGP | 2.76 | −0.54 | −0.49 | 2.73 |
| GI | High | Low | High | High |
| BBBP | Yes | No | No | Yes |
| vLROF | 0 | 0 | 0 | 0 |
| vGR | 1 | 0 | 0 | 0 |
| vVR | 0 | 0 | 0 | 0 |
| BS | 0.55 | 0.55 | 0.55 | 0.55 |
| SA | 1.00 | 3.84 | 2.00 | 2.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Debnath, S.; Debnath, B.; Debnath, P. Carvacrol: A PLpro Inhibitor of SARS-CoV-2 Is a Natural Weapon for COVID-19. Chem. Proc. 2022, 12, 11. https://doi.org/10.3390/ecsoc-26-13679
Debnath S, Debnath B, Debnath P. Carvacrol: A PLpro Inhibitor of SARS-CoV-2 Is a Natural Weapon for COVID-19. Chemistry Proceedings. 2022; 12(1):11. https://doi.org/10.3390/ecsoc-26-13679
Chicago/Turabian StyleDebnath, Sudhan, Bimal Debnath, and Pradip Debnath. 2022. "Carvacrol: A PLpro Inhibitor of SARS-CoV-2 Is a Natural Weapon for COVID-19" Chemistry Proceedings 12, no. 1: 11. https://doi.org/10.3390/ecsoc-26-13679
APA StyleDebnath, S., Debnath, B., & Debnath, P. (2022). Carvacrol: A PLpro Inhibitor of SARS-CoV-2 Is a Natural Weapon for COVID-19. Chemistry Proceedings, 12(1), 11. https://doi.org/10.3390/ecsoc-26-13679






