Deciphering the Neurosensory Olfactory Pathway and Associated Neo-Immunometabolic Vulnerabilities Implicated in COVID-Associated Mucormycosis (CAM) and COVID-19 in a Diabetes Backdrop—A Novel Perspective
Abstract
:1. Introduction
1.1. Current Taxonomy of Mucorales and Congeners
1.2. COVID-Associated Mucormycosis (CAM)
1.3. Mucormycosis-Associated Diabetes (MAD)
1.4. Immuno-Pathobiology of CAM and the Interface between COVID-19 and Mucor
1.5. The Complex Interplay of Various Factors: Mucosal Proteases and Iron Redox Stress
2. Potential Immuno-Metabolic Vulnerabilities That Can Prefigure COVID-19 and CAM-Synergistic Action of Diabetes-Associated Proteolytic and Metabolic Stress
2.1. Expanding the MSAI-Proteolytic Stress as a New Player in COVID-19 Arena
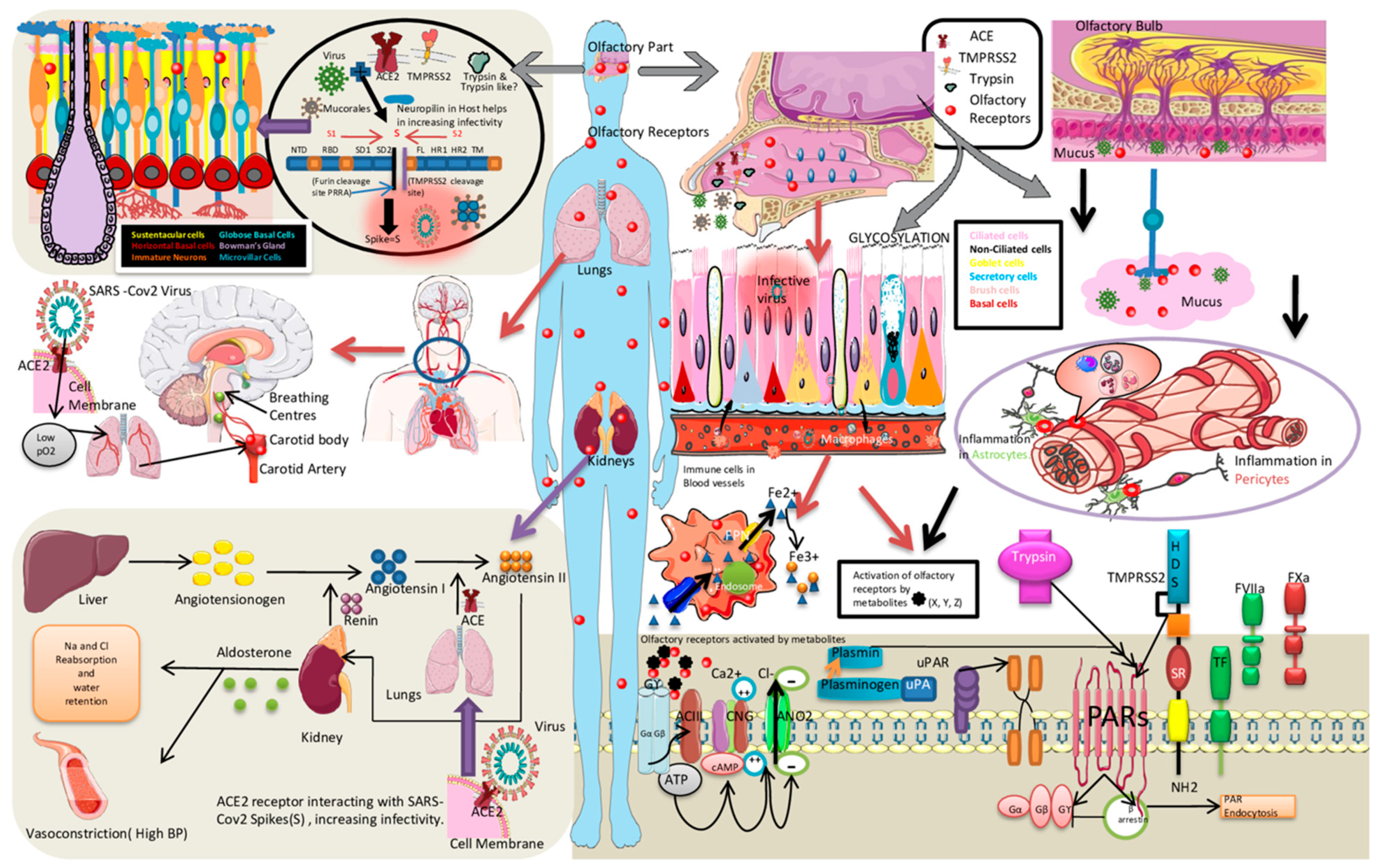
2.2. Enhanced SARS-CoV-2 Transmissibility and Role of Olfactory Mucosal Proteases
2.3. Neuro-Vascular Olfactory Mucosal Niche in Diabetes and SARS-CoV-2 Pathogenesis
2.4. Proteases as Signal Transducers—A Role beyond Spike Clipping
2.5. In Vitro Disease Modelling to Recapitulate COVID-19 and Diabetic Pathways
3. Methods
3.1. In Vitro Virus-Free Model-Establishment and Characterization of a Novel Proteolytically Tunable Plasma Based Cellular Stress Model for COVID-19 Modeling
3.2. Transcriptomic Profiling
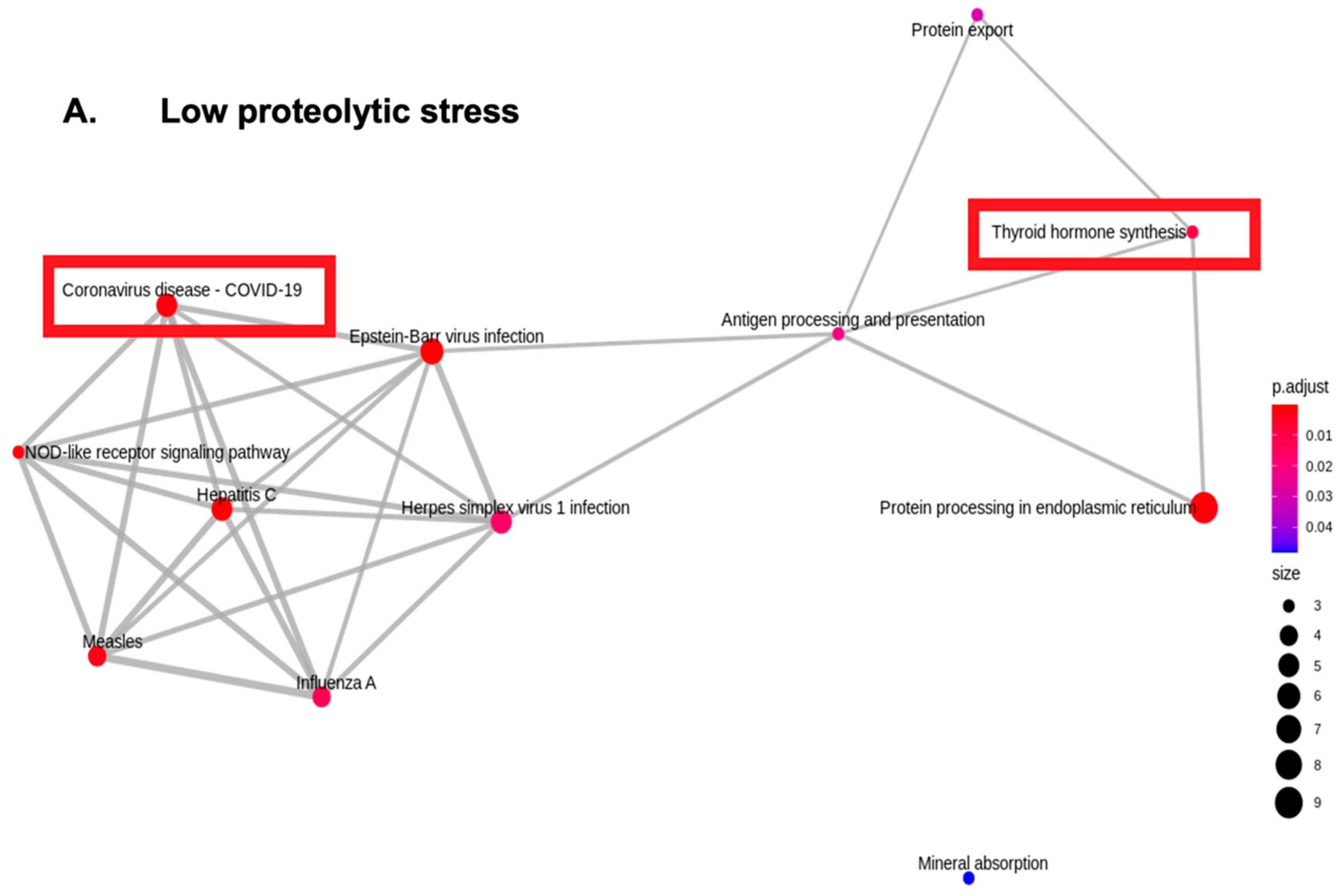
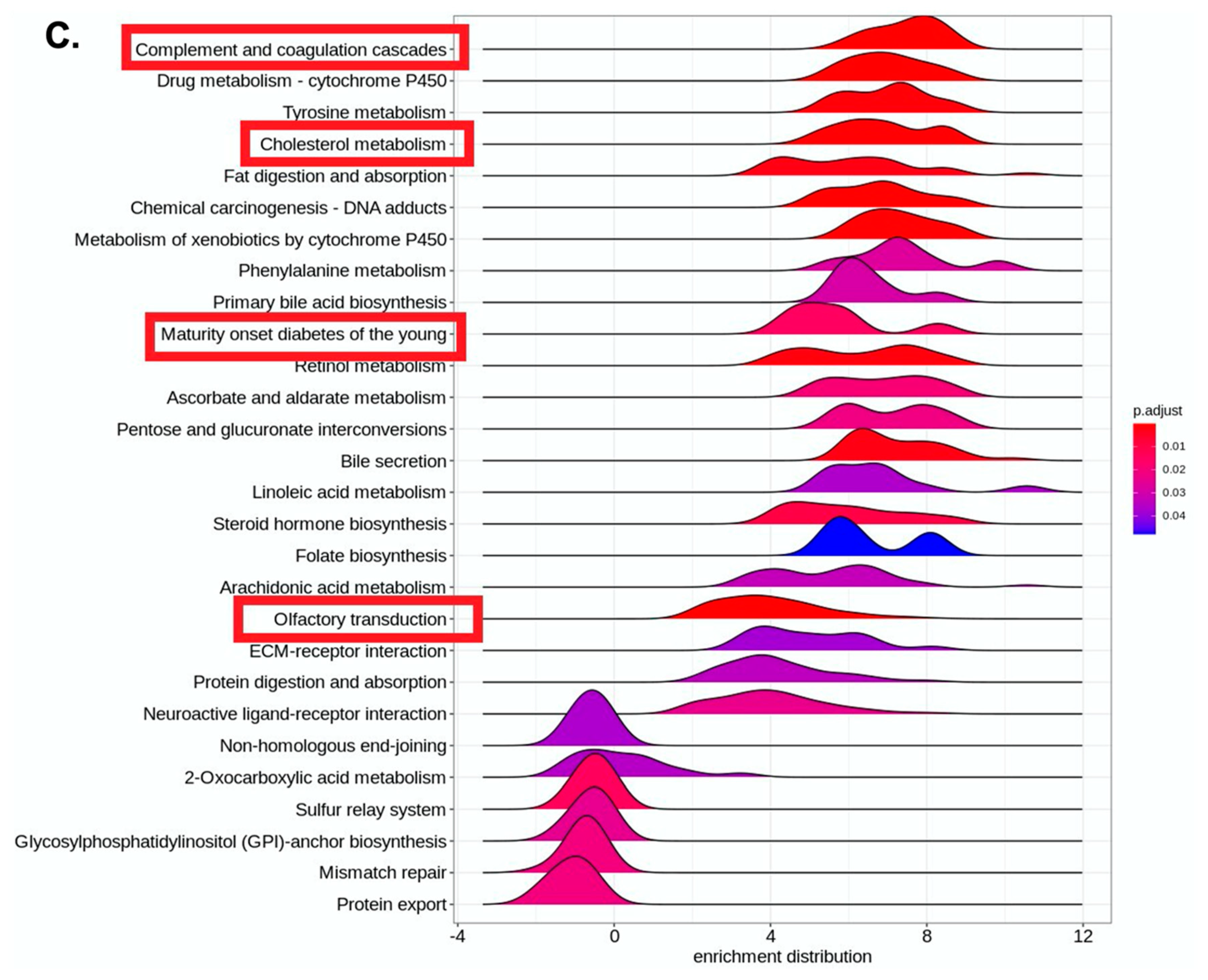
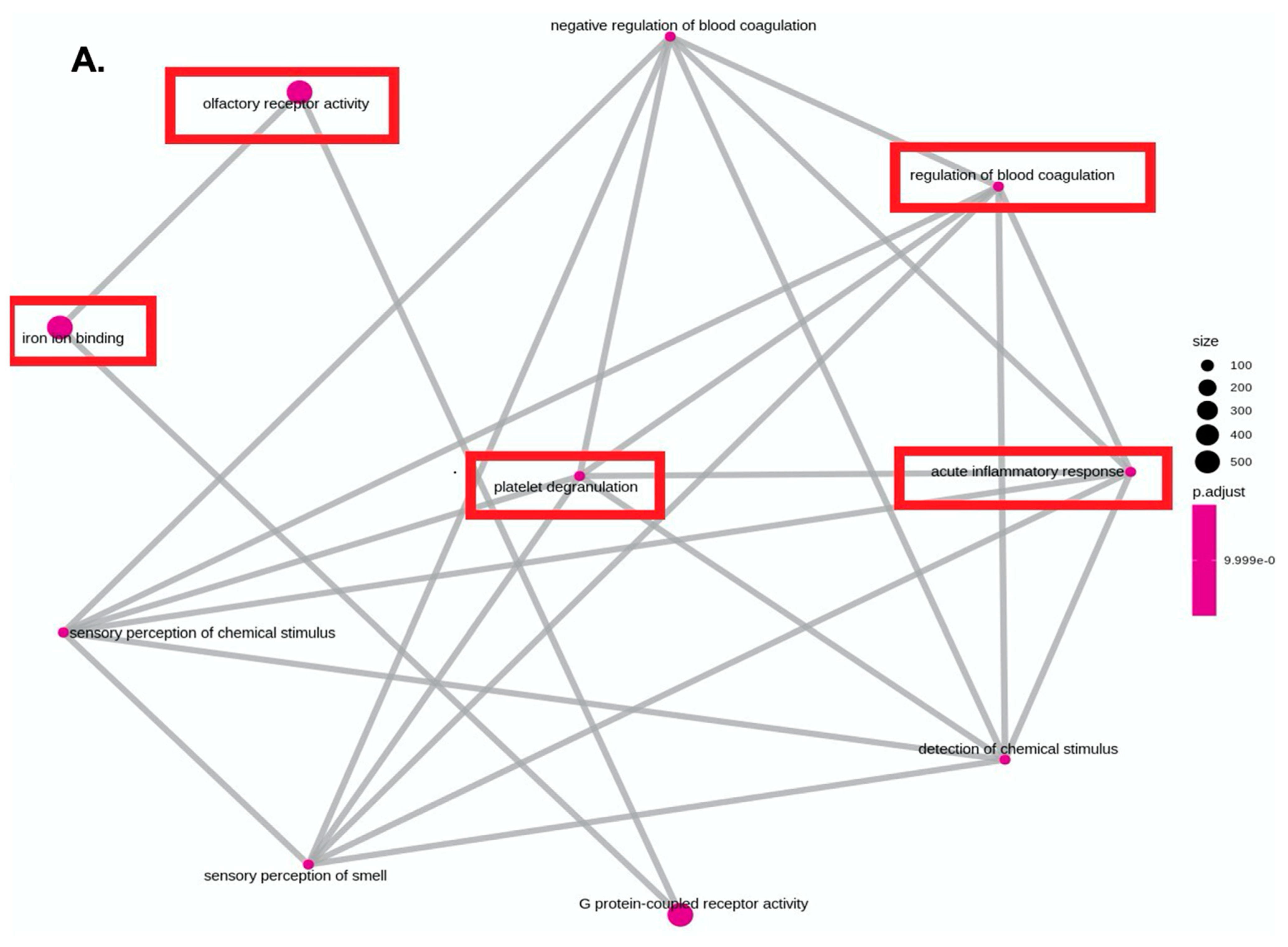
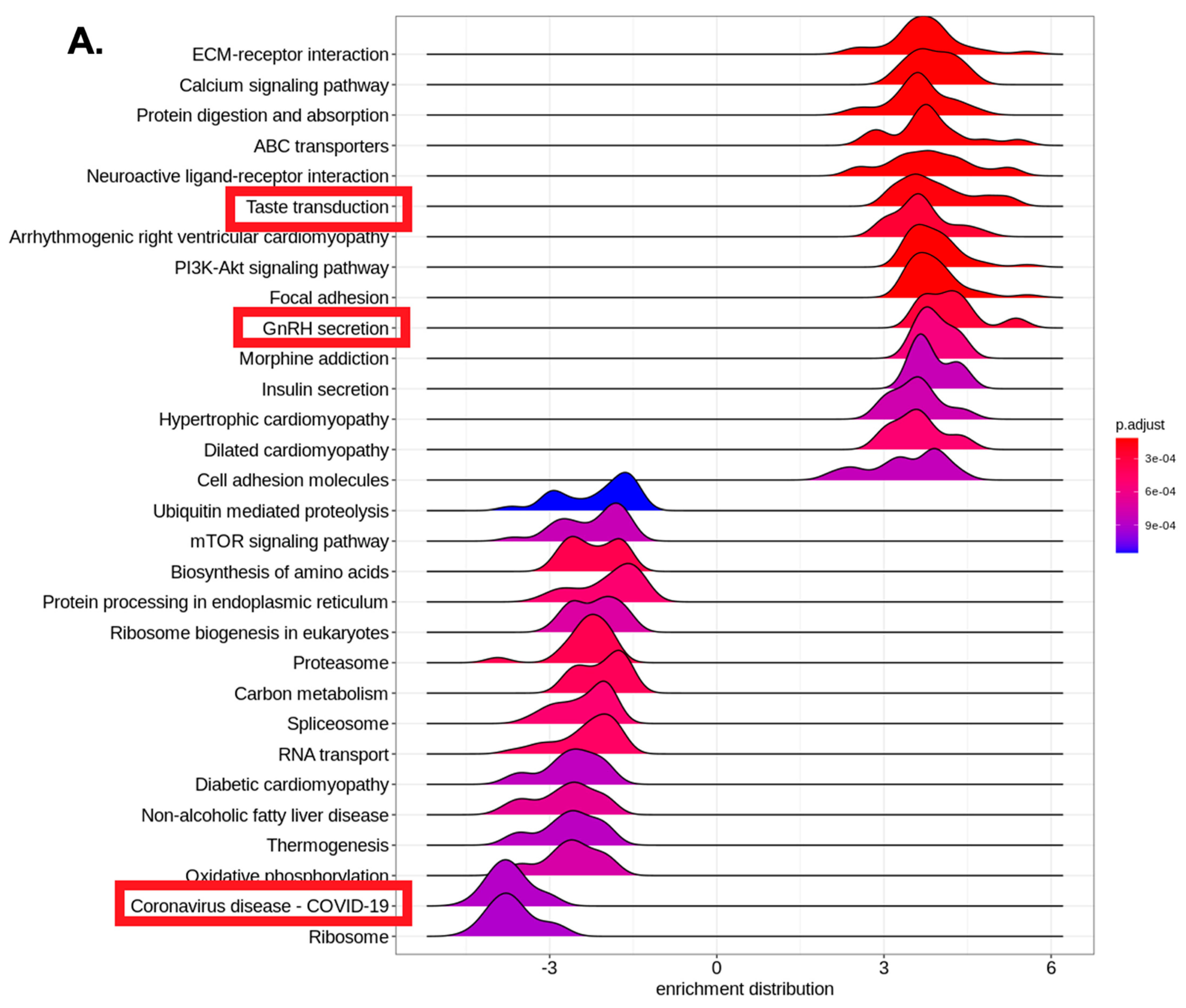
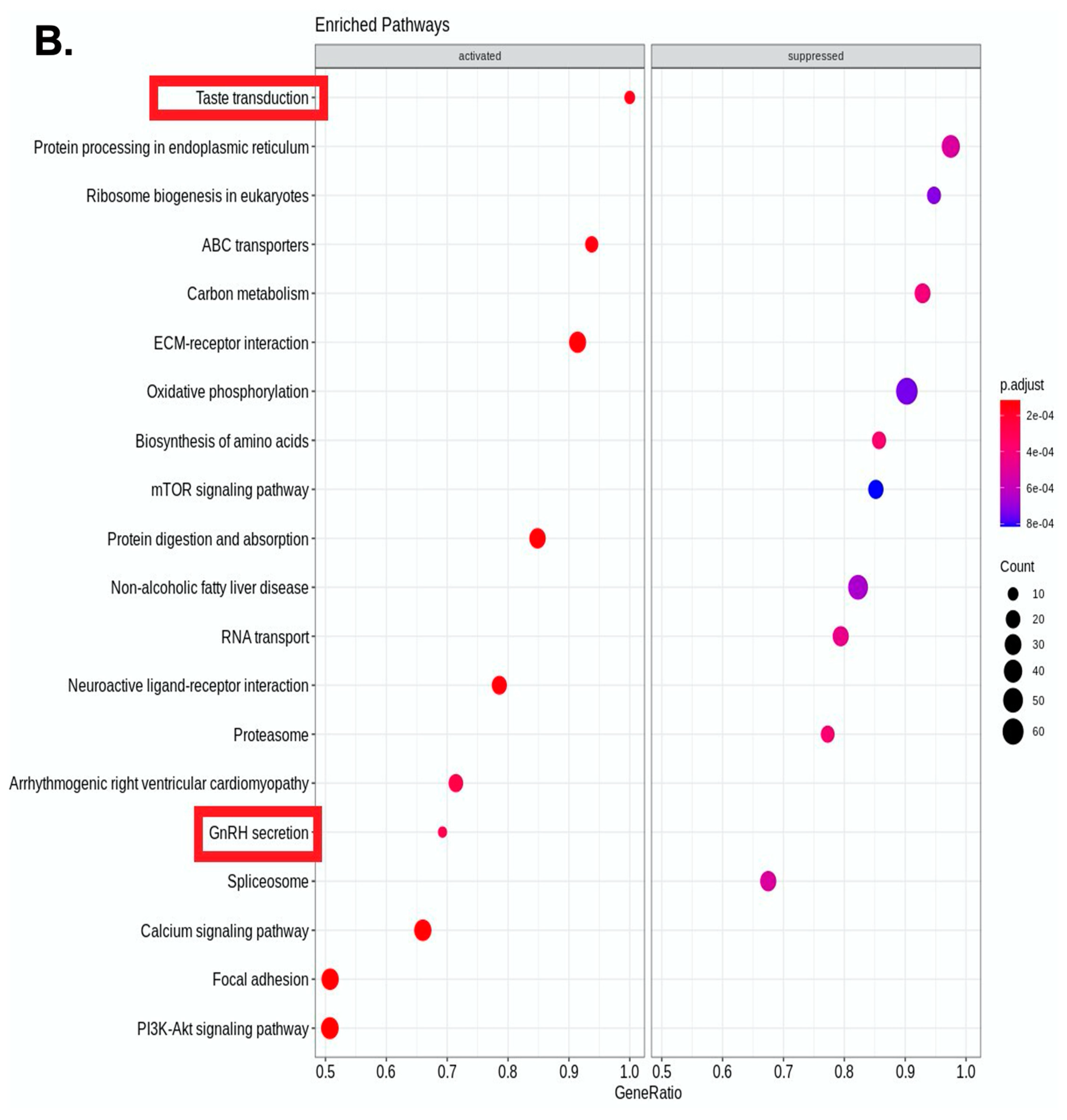
4. Results
5. Discussion
5.1. Hexosamine Biosynthetic Pathway of Glycosylation and Metabolic Stress Calibration in Olfactory Mucosa
5.2. Diabetes and Metabolic Iron Redox-Stress- Macrophages as Ferrostats
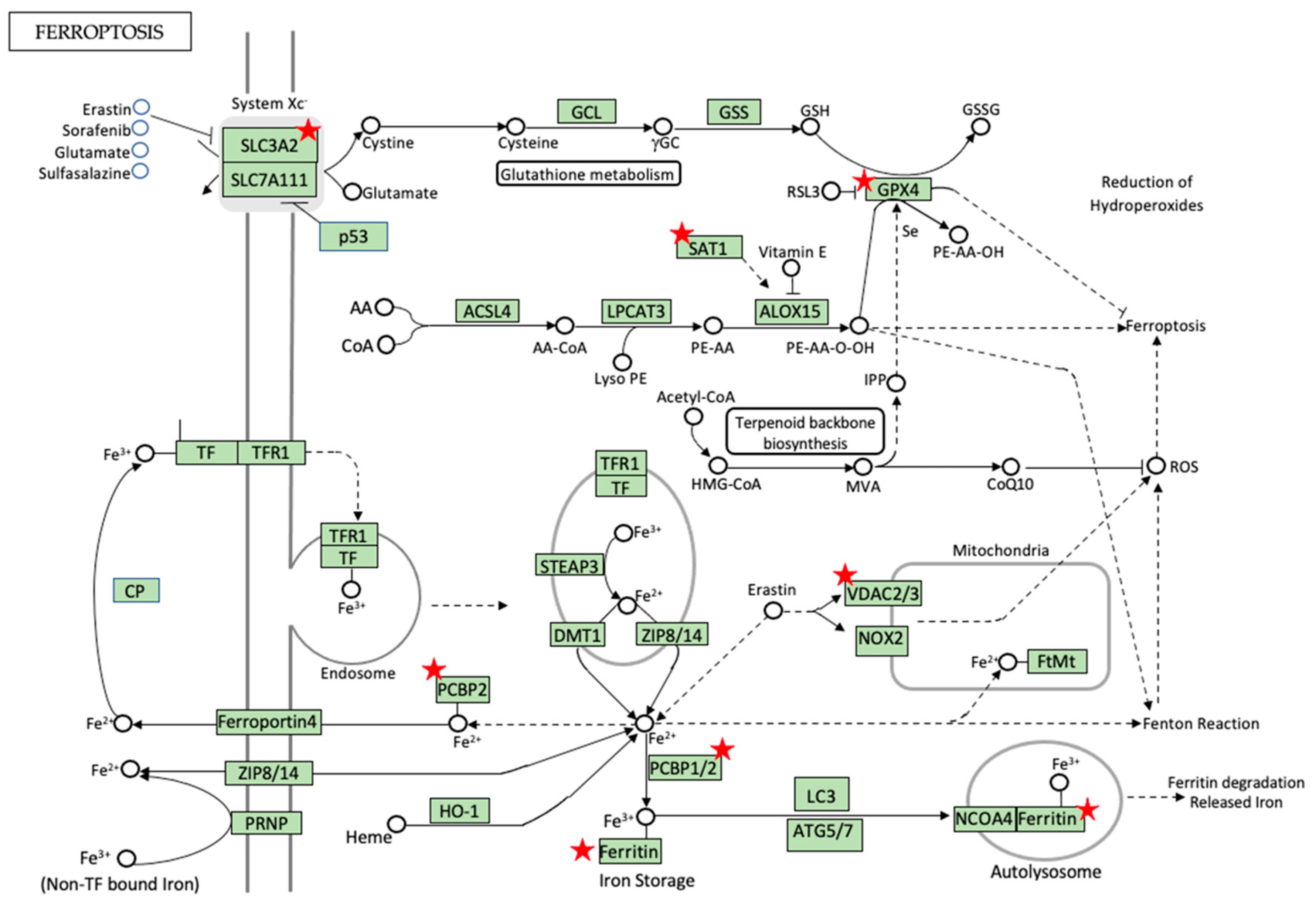
5.3. Iron Metabolism and Homeostasis—Can Ferroptosis Be the Game Changer?
5.4. Pro-Ferroptotic Labile Iron Pool (LIP) and RNA-Binding Proteins (RBPs) in Regulation of Ferroptosis and Diabetes
6. Routes of Infection and Current Perspectives in Clinical Presentation and Diagnosis
Clinical Perspective of Rhino-Orbital Mucormycosis and Patient Management
- Continued treatment of the primary immune deficiency condition.
- Correction of biochemical parameters and management of associated diabetic status.
- Aggressive debridement of the necrotic tissues of the rhino-orbital-facial region with the aim to clear all necrotic tissue and osteomyelitic bones to the maximum extent until the tissues bleed, with caution used in case of cerebral involvement to not to debride brain tissue.
- Post-debridement wound and cavity local care and adjuvant medical management with amphotericin B and/or posaconazole and isavucanazole.
- Continuation of cavity care after the completion of therapy with the regular clearance of crusts and saline irrigations for 3–6 months after treatment.
- Prosthetic rehabilitation and/or reconstruction of the defect.
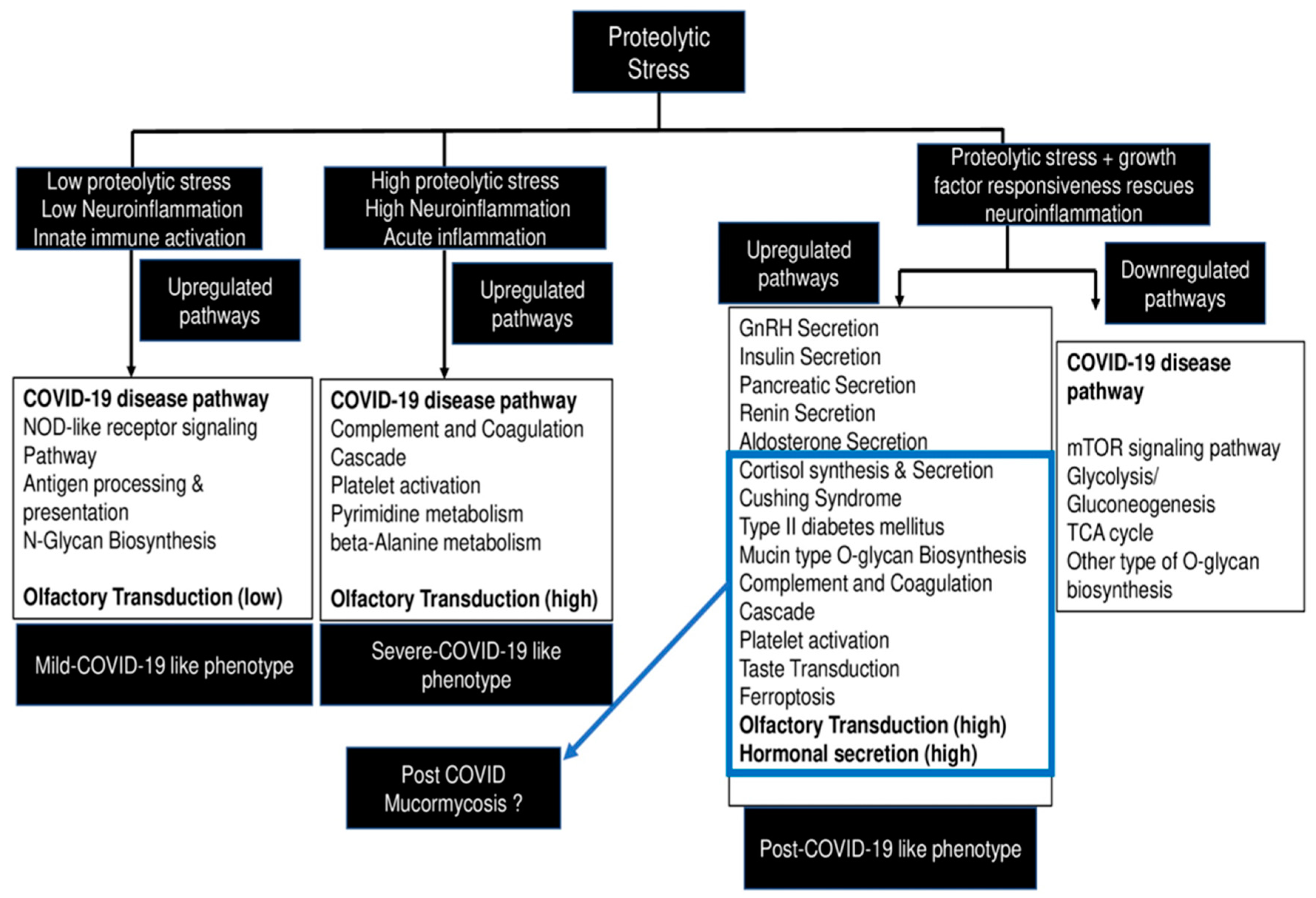
7. Future Perspective—A Working Hypothesis to Explain Development of COVID-19 and CAM in the Backdrop of Diabetes
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin-Converting Enzyme 2 |
| AMP | Adenosine Monophosphate |
| AMPK | AMP-Activated Protein Kinase |
| BAL | Bronchoalveolar Lavage |
| BDG | β-D-Glucan |
| BiPs | Binding Proteins |
| CAM | COVID-19-Associated Mucormycosis |
| CAPA | COVID-19-Associated Pulmonary Aspergillosis |
| CDC | Centers for Disease Control |
| CotH | Coat Protein Homolog |
| COVID-19 | Coronavirus Disease 2019 |
| CS | Cell Surface |
| CT | Computed Tomography |
| DAMPs | Damage-Associated Molecular Pattern |
| 2DG | 2-Deoxy-D-Glucose |
| DHPS | Deoxyhypusine Synthase |
| DKA | Diabetic keto acidosis |
| DM | Diabetes Mellitus |
| DMT1 | Divalent Metal Ion Transporter 1 |
| DOHH | Deoxyhypusine Hydroxylase |
| DPP4 | Dipeptidyl-peptidase 4 |
| eIF5a | Eukaryotic Translation Initiation Factor 5 a |
| ELISpot | Enzyme-Linked Immunospot |
| EMT | Epithelial–Mesenchymal Transition |
| EORTC | European Organization for Research and Treatment of Cancer |
| FAK | Focal Adhesion Kinase |
| Fe–GSH | Iron Glutathione |
| Fe–S | Iron Sulfur |
| FFA | Free Fatty Acids |
| FISF | Fungal Infection Study Forum |
| GM | Galactomannan |
| GnRH | Gonadotrophin-Releasing Hormone |
| GPI | Glycosylphosphatidylinositol |
| GRP78 | Glucose-regulated proteins 78 |
| HBP | Hexosamine Biosynthetic Pathway |
| HPS | High Proteolytic Stress |
| HR | High-Resolution |
| HSP70 | Heat Shock Protein 70 |
| HSPGs | Heparan Sulfate Proteoglycans |
| IC | Immunocompromised |
| IDRs | Intrinsically Disordered Regions |
| IFD | Invasive Fungal Disease |
| IFN-γ | Interferon Gamma |
| IL-1 | Interleukin 1 |
| IL-10 | Interleukin 10 |
| IL-1β | Interleukin 1 Beta |
| Il-6 | Interleukin 6 |
| IL-8 | Interleukin 8 |
| IL4 | Interleukin 4 |
| IM | Invasive Mucormycosis |
| IPA | Invasive Pulmonary Aspergillosis |
| ISHAM | International Society of Human and Animal Mycology |
| ISR | Integrated Stress Response |
| ITS Sequencing | Internal Transcribed Spacer |
| ITS | Internal Transcribed Spacer |
| KOH Mount | Potassium Hydroxide Mount |
| LIP | Labile Iron Pool |
| LMIC | Low- and Middle-Income Countries |
| LPS | Low Proteolytic Stress |
| MAD | Mucormycosis-Associated diabetes |
| MALDI–TOF | Matrix-Assisted Laser Desorption Ionization–Time-of-Flight Mass Spectrometry |
| MLKL | Mixed Lineage Kinase Linked Domain |
| MPI | Anti-Metabolic and Anti-Proteolytic Inhibitors |
| MRI | Magnetic Resonance Imaging |
| MSAI | Metabolic-Stress-associated Interactome |
| MSG | European Confederation of Medical Mycology (ECMM), and Mycoses Study Group |
| MTOR | Mammalian Target of Rapamycin |
| NBTI | Non Transferrin Bound Iron |
| NCOA4 | Nuclear Receptor Coactivator 4 |
| NONS | Nitric Oxide Nasal Spray |
| NRP1 | Neuropilin-1 |
| OE | Olfactory Epithelium |
| OSNs | Olfactory Sensory Neuron |
| PAR-2 | Protease-Activated Receptor-2 |
| PCBP | Poly(rC) Binding Protein |
| PHD | Prolyl Hydroxylase |
| PITTR | Protease-Induced Transcriptomic/Epi-Transcriptomic Reshaping |
| PM | Pulmonary Mucormycosis |
| PMNs | Polymorphoneutrophils |
| PRRs | Pattern Recognizing Receptors |
| PRSS8 | Serine Protease-8 |
| PUFAs | Plasma Membrane Unsaturated Lipids |
| qPCR | Quantitative Polymerase Chain Reaction |
| RBP | Receptor Binding Protein |
| RBPs | RNA-Binding Proteins |
| RCD | Regulated Cell Death |
| RFLP | Restriction Fragment Length Polymorphism |
| RNAi | RNAInterference |
| ROCM | Rhino-Orbital-Cerebral-Mucormycosis |
| ROS | Reactive Oxygen Species |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| SAT1 | Spermidine N1 Acetyltransferase |
| SC | Systemic Corticosteroids |
| SNCs | Sustentacular Cells |
| SPs | Serine Proteases |
| T2DM | Type 2 Diabetes Mellitus |
| TAT | Turnaround-Time |
| TCR | T-Cell Receptor |
| TLR4–IRF5 | Toll-Like Receptor 4–Activated Interferon Regulatory Factor 5 |
| TLRs | Toll-Like-Receptors |
| TMPRSS2 | Transmembrane Serine Protease 2 |
| TNF α | Tumor Necrosis Factor α |
| UDP | Uridine Diphosphate |
| VWF | Willebrand Factor |
References
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, H.; Chakrabarti, A. Global Epidemiology of Mucormycosis. J. Fungi 2019, 5, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales-Franco, B.; Nava-Villalba, M.; Medina-Guerrero, E.O.; Sánchez-Nuño, Y.A.; Davila-Villa, P.; Anaya-Ambriz, E.J.; Charles-Niño, C.L. Host-Pathogen Molecular Factors Contribute to the Pathogenesis of Rhizopus spp. in Diabetes Mellitus. Curr. Trop. Med. Rep. 2021, 8, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Ribes, J.A.; Vanover-Sams, C.L.; Baker, D.J. Zygomycetes in Human Disease. Clin. Microbiol. Rev. 2000, 13, 236–301. [Google Scholar] [CrossRef]
- Hallur, V.; Prakash, H.; Sable, M.; Preetam, C.; Purushotham, P.; Senapati, R.; Shankarnarayan, S.A.; Bag, N.D.; Rudramurthy, S.M. Cunninghamella arunalokei a New Species of Cunninghamella from India Causing Disease in an Immunocompetent Individual. J. Fungi 2021, 7, 670. [Google Scholar] [CrossRef]
- Muthu, V.; Rudramurthy, S.M.; Chakrabarti, A.; Agarwal, R. Epidemiology and Pathophysiology of COVID-19-Associated Mucormycosis: India versus the Rest of the World. Mycopathologia 2021, 186, 739–754. [Google Scholar] [CrossRef]
- Prakash, H.; Skiada, A.; Paul, R.; Chakrabarti, A.; Rudramurthy, S. Connecting the Dots: Interplay of Pathogenic Mechanisms between COVID-19 Disease and Mucormycosis. J. Fungi 2021, 7, 616. [Google Scholar] [CrossRef]
- Tandon, N.; Anjana, R.M.; Mohan, V.; Kaur, T.; Afshin, A.; Ong, K.; Mukhopadhyay, S.; Thomas, N.; Bhatia, E.; Krishnan, A.; et al. The increasing burden of diabetes and variations among the states of India: The Global Burden of Disease Study 1990–2016. Lancet Glob. Health 2018, 6, e1352–e1362. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, M.; Pal, R.; Bhadada, S.K. Intercepting the deadly trinity of mucormycosis, diabetes and COVID-19 in India. Postgrad. Med. J. 2021. [Google Scholar] [CrossRef]
- Pal, R.; Singh, B.; Bhadada, S.K.; Banerjee, M.; Bhogal, R.S.; Hage, N.; Kumar, A. COVID-19-associated mucormycosis: An updated systematic review of literature. Mycoses 2021, 64, 1452–1459. [Google Scholar] [CrossRef]
- Gupta, R.; Kesavadev, J.; Krishnan, G.; Agarwal, S.; Saboo, B.; Shah, M.; Mittal, A.; Durani, S.; Luthra, A.; Singhal, A.; et al. COVID-19 associated mucormycosis: A Descriptive Multisite Study from India. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 102322. [Google Scholar] [CrossRef] [PubMed]
- Rudramurthy, S.M.; Hoenigl, M.; Meis, J.F.; Cornely, O.A.; Muthu, V.; Gangneux, J.P.; Perfect, J.; Chakrabarti, A.; Isham, E.A. ECMM/ISHAM recommendations for clinical management of COVID-19 associated mucormycosis in low- and middle-income countries. Mycoses 2021, 64, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Denova-Gutiérrez, E.; Lopez-Gatell, H.; Alomia-Zegarra, J.L.; López-Ridaura, R.; Zaragoza-Jimenez, C.A.; Dyer-Leal, D.D.; Cortés-Alcala, R.; Villa-Reyes, T.; Gutiérrez-Vargas, R.; Rodríguez-González, K.; et al. The Association of Obesity, Type 2 Diabetes, and Hypertension with Severe Coronavirus Disease 2019 on Admission Among Mexican Patients. Obesity 2020, 28, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Heaney, A.I.; Griffin, G.D.; Simon, E.L. Newly diagnosed diabetes and diabetic ketoacidosis precipitated by COVID-19 infection. Am. J. Emerg. Med. 2020, 38, 2491.e3–2491.e4. [Google Scholar] [CrossRef] [PubMed]
- Alekseyev, K.; Didenko, L.; Chaudhry, B. Rhinocerebral Mucormycosis and COVID-19 Pneumonia. J. Med. Cases 2021, 12, 85–89. [Google Scholar] [CrossRef]
- John, T.; Jacob, C.; Kontoyiannis, D. When Uncontrolled Diabetes Mellitus and Severe COVID-19 Converge: The Perfect Storm for Mucormycosis. J. Fungi 2021, 7, 298. [Google Scholar] [CrossRef]
- Ayelign, B.; Negash, M.; Genetu, M.; Wondmagegn, T.; Shibabaw, T. Immunological Impacts of Diabetes on the Susceptibility of Mycobacterium tuberculosis. J. Immunol. Res. 2019, 2019, 6196532. [Google Scholar] [CrossRef]
- Affinati, A.H.; Wallia, A.; Gianchandani, R.Y. Severe hyperglycemia and insulin resistance in patients with SARS-CoV-2 infection: A report of two cases. Clin. Diabetes Endocrinol. 2021, 7, 8. [Google Scholar] [CrossRef]
- Prakash, H.; Chakrabarti, A. Epidemiology of Mucormycosis in India. Microorganisms 2021, 9, 523. [Google Scholar] [CrossRef]
- Garg, D.; Muthu, V.; Sehgal, I.S.; Ramachandran, R.; Kaur, H.; Bhalla, A.; Puri, G.D.; Chakrabarti, A.; Agarwal, R. Coronavirus Disease (COVID-19) Associated Mucormycosis (CAM): Case Report and Systematic Review of Literature. Mycopathologia 2021, 186, 289–298. [Google Scholar] [CrossRef]
- Alfishawy, M.; Elbendary, A.; Younes, A.; Negm, A.; Hassan, W.S.; Osman, S.H.; Nassar, M.; Elanany, M.G. Diabetes mellitus and Coronavirus Disease (COVID-19) Associated Mucormycosis (CAM): A wake-up call from Egypt. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 102195. [Google Scholar] [CrossRef] [PubMed]
- Farias, L.A.B.G.; Damasceno, L.S.; Bandeira, S.P.; Barreto, F.K.D.A.; Leitão, T.D.M.J.S.; Cavalcanti, L.P.D.G. COVID-19 associated Mucormycosis (CAM): Should Brazil be on alert? Rev. Soc. Bras. Med. Trop. 2021, 54, e0410-2021. [Google Scholar] [CrossRef] [PubMed]
- Epidemiological Alert: COVID-19 Associated Mucormycosis (11 June 2021). Available online: https://iris.paho.org/handle/10665.2/54284 (accessed on 11 January 2022).
- Hoenigl, M.; Seidel, D.; Carvalho, A.; Rudramurthy, S.M.; Arastehfar, A.; Gangneux, J.P.; Nasir, N.; Bonifaz, A.; Araiza, J.; Klimko, N.; et al. The Emergence of COVID-19 Associated Mucormycosis: Analysis of Cases from 18 Countries. Lancet Microbe. 2022. [Google Scholar] [CrossRef]
- Patel, A.; Agarwal, R.; Rudramurthy, S.M.; Shevkani, M.; Xess, I.; Sharma, R.; Savio, J.; Sethuraman, N.; Madan, S.; Shastri, P.; et al. Multicenter Epidemiologic Study of Coronavirus Disease–Associated Mucormycosis, India. Emerg. Infect. Dis. 2021, 27, 2349–2359. [Google Scholar] [CrossRef]
- Banerjee, I.; Robinson, J.; Asim, M.; Sathian, B.; Banerjee, I. Mucormycosis and COVID-19 an epidemic in a pandemic? Nepal J. Epidemiol. 2021, 11, 1034–1039. [Google Scholar] [CrossRef]
- Statement from Health Minister, Government of India to Press. Available online: https://www.tribuneindia.com/news/nation/28-252-Black-fungus-cases-in-india-265262 (accessed on 11 January 2022).
- Cornely, O.A.; Alastruey-Izquierdo, A.; Arenz, D.; Chen, S.C.A.; Dannaoui, E.; Hochhegger, B.; Hoenigl, M.; Jensen, H.E.; Lagrou, K.; Lewis, R.E.; et al. Global guideline for the diagnosis and management of mucormycosis: An initiative of the European Confederation of Medical Mycology in cooperation with the Mycoses Study Group Education and Research Consortium. Lancet Infect. Dis. 2019, 19, e405–e421. [Google Scholar] [CrossRef]
- Farmakiotis, D.; Kontoyiannis, D.P. Mucormycoses. Infect. Dis. Clin. N. Am. 2016, 30, 143–163. [Google Scholar] [CrossRef]
- Sabirli, R.; Koseler, A.; Goren, T.; Turkcuer, I.; Kurt, O. High GRP78 levels in COVID-19 infection: A case-control study. Life Sci. 2021, 265, 118781. [Google Scholar] [CrossRef]
- Rayner, J.O.; Roberts, R.A.; Kim, J.; Poklepovic, A.; Roberts, J.L.; Booth, L.; Dent, P. AR12 (OSU-03012) suppresses GRP78 expression and inhibits SARS-CoV-2 replication. Biochem. Pharmacol. 2020, 182, 114227. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Sarma, P.; Sharma, D.J.; Das, K.K.; Kaur, H.; Prajapat, M.; Kumar, S.; Bansal, S.; Prakash, A.; Avti, P.; et al. Rhino-Orbital-Cerebral-Mucormycosis in COVID-19: A Systematic Review. Indian J. Pharmacol. 2021, 53, 317–327. [Google Scholar] [CrossRef]
- Devana, S.K.; Gupta, V.G.; Mavuduru, R.S.; Bora, G.S.; Sharma, A.P.; Parmar, K.M.; Kumar, S.; Mete, U.K.; Singh, S.K.; Mandal, A.K.; et al. Isolated Renal Mucormycosis in Immunocompetent Hosts: Clinical Spectrum and Management Approach. Am. J. Trop. Med. Hyg. 2019, 100, 791–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, N.V.S.; Natti, R.S.; Radha, T.; Sharma, M.; Chintham, M. Skull Base Mucormycosis in an Immunocompetent Patient: A Case Report and Literature Review. Indian J. Otolaryngol. Head Neck Surg. 2018, 71, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Walther, G.; Wagner, L.; Kurzai, O. Updates on the Taxonomy of Mucorales with an Emphasis on Clinically Important Taxa. J. Fungi 2019, 5, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, S.M.S.; Borghei, P. Rhinocerebral mucormycosis: Pathways of spread. Eur. Arch. Oto-Rhino-Laryngol. Head Neck 2005, 262, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Zhang, Y.; Lee, A.S. Beyond the endoplasmic reticulum: Atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem. J. 2011, 434, 181–188. [Google Scholar] [CrossRef]
- Kwon, J.-W.; Jung, I.; Jee, D. Glucose-regulated protein 78 in the aqueous humor in diabetic macular edema patients. Medicine 2018, 97, e12757. [Google Scholar] [CrossRef]
- Liu, M.; Spellberg, B.; Phan, Q.T.; Fu, Y.; Fu, Y.; Lee, A.; Edwards, J.E.; Filler, S.G.; Ibrahim, A.S. The endothelial cell receptor GRP78 is required for mucormycosis pathogenesis in diabetic mice. J. Clin. Investig. 2010, 120, 1914–1924. [Google Scholar] [CrossRef] [Green Version]
- Ha, D.P.; Van Krieken, R.; Carlos, A.J.; Lee, A.S. The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J. Infect. 2020, 81, 452–482. [Google Scholar] [CrossRef]
- Nasir, N.; Sayeed, M.A.; Jamil, B. Ralstonia pickettii Bacteremia: An Emerging Infection in a Tertiary Care Hospital Setting. Cureus 2019, 11, e5084. [Google Scholar] [CrossRef] [Green Version]
- Dolatabadi, S.; Scherlach, K.; Figge, M.; Hertweck, C.; Dijksterhuis, J.; Menken, S.B.; De Hoog, G.S. Food preparation with mucoralean fungi: A potential biosafety issue? Fungal Biol. 2016, 120, 393–401. [Google Scholar] [CrossRef]
- Rickerts, V.; Böhme, A.; Viertel, A.; Behrendt, G.; Jacobi, V.; Tintelnot, K.; Just-Nübling, G. Cluster of Pulmonary Infections Caused byCunninghamella bertholletiaein Immunocompromised Patients. Clin. Infect. Dis. 2000, 31, 910–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lax, C.; Pérez-Arques, C.; Navarro-Mendoza, M.I.; Cánovas-Márquez, J.T.; Tahiri, G.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Murcia-Flores, L.; Navarro, E.; Garre, V.; et al. Genes, Pathways, and Mechanisms Involved in the Virulence of Mucorales. Genes 2020, 11, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trieu, T.A.; Navarro-Mendoza, M.I.; Perez-Arques, C.; Sanchis, M.; Capilla, J.; Navarro-Rodríguez, P.; Lopez-Fernandez, L.; Torres-Martínez, S.; Garre, V.; Ruiz-Vázquez, R.M.; et al. RNAi-Based Functional Genomics Identifies New Virulence Determinants in Mucormycosis. PLoS Pathog. 2017, 13, e1006150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirakawa, J. Pancreatic β-cell fate in subjects with COVID-19. J. Diabetes Investig. 2021, 12, 2126–2128. [Google Scholar] [CrossRef]
- Wu, C.-T.; Lidsky, P.V.; Xiao, Y.; Lee, I.T.; Cheng, R.; Nakayama, T.; Jiang, S.; Demeter, J.; Bevacqua, R.J.; Chang, C.A.; et al. SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metab. 2021, 33, 1565–1576.e5. [Google Scholar] [CrossRef]
- Tang, X.; Uhl, S.; Zhang, T.; Xue, D.; Li, B.; Vandana, J.J.; Acklin, J.A.; Bonnycastle, L.L.; Narisu, N.; Erdos, M.R.; et al. SARS-CoV-2 infection induces beta cell transdifferentiation. Cell Metab. 2021, 33, 1577–1591.e7. [Google Scholar] [CrossRef]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [Green Version]
- Menachery, V.D.; Dinnon, K.H.; Yount, B.L.; McAnarney, E.T.; Gralinski, L.E.; Hale, A.; Graham, R.L.; Scobey, T.; Anthony, S.J.; Wang, L.; et al. Trypsin Treatment Unlocks Barrier for Zoonotic Bat Coronavirus Infection. J. Virol. 2020, 94, e01774-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Van Der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021, 6, 1219–1232. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; St Denis, K.J.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, H.; Tebbutt, S.J. Long-term effects on survivors with COVID-19. Lancet 2021, 398, 1872. [Google Scholar] [CrossRef]
- Khan, M.; Yoo, S.-J.; Clijsters, M.; Backaert, W.; Vanstapel, A.; Speleman, K.; Lietaer, C.; Choi, S.; Hether, T.D.; Marcelis, L.; et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 2021, 184, 5932–5949.e15. [Google Scholar] [CrossRef]
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brünink, S.; Greuel, S.; et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175. [Google Scholar] [CrossRef] [PubMed]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19–related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sievert, D.; Clark, A.E.; Lee, S.; Federman, H.; Gastfriend, B.D.; Shusta, E.V.; Palecek, S.P.; Carlin, A.F.; Gleeson, J.G. A human three-dimensional neural-perivascular ‘assembloid’ promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nat. Med. 2021, 27, 1600–1606. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Kim, J.; Hong, S.P.; Choi, S.Y.; Yang, M.J.; Ju, Y.S.; Kim, Y.T.; Kim, H.M.; Rahman, T.; Chung, M.K.; et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Investig. 2021, 131, e148517. [Google Scholar] [CrossRef]
- Lechien, J.R.; Chiesa-Estomba, C.M.; Beckers, E.; Mustin, V.; Ducarme, M.; Journe, F.; Marchant, A.; Jouffe, L.; Barillari, M.R.; Cammaroto, G.; et al. Prevalence and 6-month recovery of olfactory dysfunction: A multicentre study of 1363 COVID-19 patients. J. Intern. Med. 2021, 290, 451–461. [Google Scholar] [CrossRef]
- Vaira, L.A.; Deiana, G.; Lechien, J.R.; De Vito, A.; Cossu, A.; Dettori, M.; Del Rio, A.; Saussez, S.; Madeddu, G.; Babudieri, S.; et al. Correlations Between Olfactory Psychophysical Scores and SARS-CoV-2 Viral Load in COVID-19 Patients. Laryngoscope 2021, 131, 2312–2318. [Google Scholar] [CrossRef]
- Xydakis, M.S.; Albers, M.W.; Holbrook, E.H.; Lyon, D.M.; Shih, R.Y.; Frasnelli, J.A.; Pagenstecher, A.; Kupke, A.; Enquist, L.W.; Perlman, S. Post-viral effects of COVID-19 in the olfactory system and their implications. Lancet Neurol. 2021, 20, 753–761. [Google Scholar] [CrossRef]
- Butowt, R.; Meunier, N.; Bryche, B.; von Bartheld, C.S. The olfactory nerve is not a likely route to brain infection in COVID-19: A critical review of data from humans and animal models. Acta Neuropathol. 2021, 141, 809–822. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van Den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.-J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef]
- Fodoulian, L.; Tuberosa, J.; Rossier, D.; Boillat, M.; Kan, C.; Pauli, V.; Egervari, K.; Lobrinus, J.A.; Landis, B.N.; Carleton, A.; et al. SARS-CoV-2 Receptor and Entry Genes Are Expressed by Sustentacular Cells in the Human Olfactory Neuroepithelium. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Cooper, K.; Brann, D.H.; Farruggia, M.C.; Bhutani, S.; Pellegrino, R.; Tsukahara, T.; Weinreb, C.; Joseph, P.V.; Larson, E.D.; Parma, V.; et al. COVID-19 and the Chemical Senses: Supporting Players Take Center Stage. Neuron 2020, 107, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, C.; Blanchard, K.; Bacigalupo, J.; Vergara, C. Possible ATP trafficking by ATP-shuttles in the olfactory cilia and glucose transfer across the olfactory mucosa. FEBS Lett. 2019, 593, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Villar, P.S.; Delgado, R.; Vergara, C.; Reyes, J.G.; Bacigalupo, J. Energy Requirements of Odor Transduction in the Chemosensory Cilia of Olfactory Sensory Neurons Rely on Oxidative Phosphorylation and Glycolytic Processing of Extracellular Glucose. J. Neurosci. 2017, 37, 5736–5743. [Google Scholar] [CrossRef] [Green Version]
- Liang, F. Olfactory receptor neuronal dendrites become mostly intra-sustentacularly enwrapped upon maturity. J. Anat. 2018, 232, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Fengyi, L. Sustentacular Cell Enwrapment of Olfactory Receptor Neuronal Dendrites: An Update. Genes 2020, 11, 493. [Google Scholar] [CrossRef] [PubMed]
- Shelton, J.F.; Shastri, A.J.; Aslibekyan, S.; Auton, A. The UGT2A1/UGT2A2 Locus Is Associated with COVID-19-Related loss of smell or taste. Nat Genet. 2022, 54, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Brazil, J.C.; Parkos, C.A. Finding the sweet spot: Glycosylation mediated regulation of intestinal inflammation. Mucosal Immunol. 2021, 15, 211–222. [Google Scholar] [CrossRef]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A Transmembrane Serine Protease Is Linked to the Severe Acute Respiratory Syndrome Coronavirus Receptor and Activates Virus Entry. J. Virol. 2010, 85, 873–882. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J.; et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020, 69, 1010–1018. [Google Scholar] [CrossRef]
- Fang, Y.; Liu, H.; Huang, H.; Li, H.; Saqi, A.; Qiang, L.; Que, J. Distinct stem/progenitor cells proliferate to regenerate the trachea, intrapulmonary airways and alveoli in COVID-19 patients. Cell Res. 2020, 30, 705–707. [Google Scholar] [CrossRef]
- Witkowski, M.; Tizian, C.; Ferreira-Gomes, M.; Niemeyer, D.; Jones, T.C.; Heinrich, F.; Frischbutter, S.; Angermair, S.; Hohnstein, T.; Mattiola, I.; et al. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature 2021, 600, 295–301. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Yamamoto, K.; Kataoka, H.; Izumi, A.; Yamashita, F.; Kiwaki, T.; Nishida, T.; Camerer, E.; Fukushima, T. Protease-activated receptor-2 accelerates intestinal tumor formation through activation of nuclear factor-κB signaling and tumor angiogenesis in Apc Min/+ mice. Cancer Sci. 2020, 111, 1193–1202. [Google Scholar] [CrossRef] [Green Version]
- Antoniak, S.; Mackman, N. Multiple roles of the coagulation protease cascade during virus infection. Blood 2014, 123, 2605–2613. [Google Scholar] [CrossRef] [Green Version]
- Nhu, Q.M.; Shirey, K.; Teijaro, J.R.; Farber, D.; Netzel-Arnett, S.; Antalis, T.M.; Fasano, A.; Vogel, S.N. Novel signaling interactions between proteinase-activated receptor 2 and Toll-like receptors in vitro and in vivo. Mucosal Immunol. 2010, 3, 29–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weithauser, A.; Rauch, U. Role of protease-activated receptors for the innate immune response of the heart. Trends Cardiovasc. Med. 2014, 24, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Rallabhandi, P.; Awomoyi, A.; Thomas, K.E.; Phalipon, A.; Fujimoto, Y.; Fukase, K.; Kusumoto, S.; Qureshi, N.; Sztein, M.B.; Vogel, S.N. Differential Activation of Human TLR4 byEscherichia coliandShigella flexneri2a Lipopolysaccharide: Combined Effects of Lipid A Acylation State and TLR4 Polymorphisms on Signaling. J. Immunol. 2008, 180, 1139–1147. [Google Scholar] [CrossRef] [Green Version]
- Antoniak, S. The coagulation system in host defense. Res. Pract. Thromb. Haemost. 2018, 2, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Wojtukiewicz, M.Z.; Hempel, D.; Sierko, E.; Tucker, S.C.; Honn, K.V. Protease-activated receptors (PARs)—biology and role in cancer invasion and metastasis. Cancer Metastasis Rev. 2015, 34, 775–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasthuri, R.S.; Taubman, M.B.; Mackman, N. Role of Tissue Factor in Cancer. J. Clin. Oncol. 2009, 27, 4834–4838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camerer, E.; Huang, W.; Coughlin, S.R. Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc. Natl. Acad. Sci. USA 2000, 97, 5255–5260. [Google Scholar] [CrossRef] [Green Version]
- Posma, J.J.; Grover, S.; Hisada, Y.; Owens, A.P.; Antoniak, S.; Spronk, H.M.; Mackman, N. Roles of Coagulation Proteases and PARs (Protease-Activated Receptors) in Mouse Models of Inflammatory Diseases. Arter. Thromb. Vasc. Biol. 2019, 39, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Bryzek, D.; Ciaston, I.; Dobosz, E.; Gasiorek, A.; Makarska, A.; Sarna, M.; Eick, S.; Puklo, M.; Lech, M.; Potempa, B.; et al. Triggering NETosis via protease-activated receptor (PAR)-2 signaling as a mechanism of hijacking neutrophils function for pathogen benefits. PLoS Pathog. 2019, 15, e1007773. [Google Scholar] [CrossRef]
- Sharma, M. Epithelial Cells Promote Fibroblast-Mediated Contraction of Collagen Gels by Secreting BFGF. In Proceedings of the Association for Research in Vision and Ophthalmology (ARVO) Annual Meeting, Seattle, WA, USA, 5–9 May 2013. [Google Scholar]
- Sharma, M. Establishment and Characterization of a Novel Serine Protease Induced Reprograming (SPIR) Method with Ap-plications in Ocular Tissue Regeneration. In Proceedings of the Association for Research in Vision and Ophthalmology (ARVO) Annual Meeting, Baltimore, MD, USA, 7–11 May 2017. [Google Scholar]
- Sharma, M.; Kumar, R.; Sharma, S.; Thomas, B.; Kapatia, G.; Singh, G.; Bal, A.; Ram, J.; Bhasin, M.; Guptasarma, P.; et al. Sus-tained Exposure to Trypsin Causes Cells to Transition into a State of Reversible Stemness That Is Amenable to Transdiffer-entiation. bioRxiv 2019, 679928. [Google Scholar] [CrossRef]
- Sharma, M.; Panda, N.K. Proteomic Profiling of Protease-Primed Virus-Permissive Caco-2 Cells Display Abor-tive-Interferon Pathway and Deregulated Thromboinflammatory SERPINS. Preprints 2020, 2020060206. [Google Scholar] [CrossRef]
- Humphries, F.; Shmuel-Galia, L.; Jiang, Z.; Wilson, R.; Landis, P.; Ng, S.-L.; Parsi, K.M.; Maehr, R.; Cruz, J.; Morales-Ramos, A.; et al. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. Sci. Immunol. 2021, 6, eabi9002. [Google Scholar] [CrossRef]
- Clinical Trails Arena. Available online: https://www.clinicaltrialsarena.com/news/sanotize-nasal-spray-reduces-covid-19-viral-load-uk-clinical-trail/ (accessed on 7 January 2022).
- Winchester, S.; John, S.; Jabbar, K.; John, I. Clinical efficacy of nitric oxide nasal spray (NONS) for the treatment of mild COVID-19 infection. J. Infect. 2021, 83, 237–279. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, R.; Joshi, S.R.; Misra, A. Mucormycosis in COVID-19: A systematic review of cases reported worldwide and in India. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 102146. [Google Scholar] [CrossRef] [PubMed]
- Revannavar, S.M.; Supriya, P.; Samaga, L.; Vineeth, V. COVID-19 triggering mucormycosis in a susceptible patient: A new phenomenon in the developing world? BMJ Case Rep. 2021, 14, e241663. [Google Scholar] [CrossRef] [PubMed]
- Daryabor, G.; Atashzar, M.R.; Kabelitz, D.; Meri, S.; Kalantar, K. The Effects of Type 2 Diabetes Mellitus on Organ Metabolism and the Immune System. Front. Immunol. 2020, 11, 1582. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, M.A.; Saavedra, V.; Mangan, M.S.J.; Penno, A.; Thiele, C.; Latz, E.; Kuerschner, L. 1-Deoxysphingolipids cause autophagosome and lysosome accumulation and trigger NLRP3 inflammasome activation. Autophagy 2020, 17, 1947–1961. [Google Scholar] [CrossRef]
- Alzaid, F.; Julla, J.; Diedisheim, M.; Potier, C.; Potier, L.; Velho, G.; Gaborit, B.; Manivet, P.; Germain, S.; Vidal-Trecan, T.; et al. Monocytopenia, monocyte morphological anomalies and hyperinflammation characterise severe COVID-19 in type 2 diabetes. EMBO Mol. Med. 2020, 12, e13038. [Google Scholar] [CrossRef]
- Al-Rashed, F.; Sindhu, S.; Arefanian, H.; Al Madhoun, A.; Kochumon, S.; Thomas, R.; Al-Kandari, S.; Alghaith, A.; Jacob, T.; Al-Mulla, F.; et al. Repetitive Intermittent Hyperglycemia Drives the M1 Polarization and Inflammatory Responses in THP-1 Macrophages Through the Mechanism Involving the TLR4-IRF5 Pathway. Cells 2020, 9, 1892. [Google Scholar] [CrossRef]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.N.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [Green Version]
- Reusch, N.; De Domenico, E.; Bonaguro, L.; Schulte-Schrepping, J.; Baßler, K.; Schultze, J.L.; Aschenbrenner, A.C. Neutrophils in COVID-19. Front. Immunol. 2021, 12, 652470. [Google Scholar] [CrossRef]
- Szabo, P.A.; Dogra, P.; Gray, J.I.; Wells, S.B.; Connors, T.J.; Weisberg, S.P.; Krupska, I.; Matsumoto, R.; Poon, M.M.; Idzikowski, E.; et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity 2021, 54, 797–814.e6. [Google Scholar] [CrossRef]
- Courjon, J.; Dufies, O.; Robert, A.; Bailly, L.; Torre, C.; Chirio, D.; Contenti, J.; Vitale, S.; Loubatier, C.; Doye, A.; et al. Heterogeneous NLRP3 inflammasome signature in circulating myeloid cells as a biomarker of COVID-19 severity. Blood Adv. 2021, 5, 1523–1534. [Google Scholar] [CrossRef]
- de Sá-Ferreira, C.O.; da Costa, C.H.M.; Guimarães, J.C.W.; Sampaio, N.S.; Silva, L.d.M.L.; de Mascarenhas, L.P.; Rodrigues, N.G.; dos Santos, T.L.; Campos, S.; Young, E.C. Diabetic ketoacidosis and COVID-19: What have we learned so far? Am. J. Physiol. Metab. 2022, 322, E44–E53. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.A.; Groß, R.; Conzelmann, C.; Krüger, J.; Merle, U.; Steinhart, J.; Weil, T.; Koepke, L.; Bozzo, C.P.; Read, C.; et al. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nat. Metab. 2021, 3, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Perico, L.; Benigni, A.; Casiraghi, F.; Ng, L.F.P.; Renia, L.; Remuzzi, G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat. Rev. Nephrol. 2020, 17, 46–64. [Google Scholar] [CrossRef]
- Silva, D.; Lima, C.; Magalhães, V.; Baltazar, L.; Peres, N.; Caligiorne, R.; Moura, A.; Fereguetti, T.; Martins, J.; Rabelo, L.; et al. Fungal and bacterial coinfections increase mortality of severely ill COVID-19 patients. J. Hosp. Infect. 2021, 113, 145–154. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Moreno-García, E.; Hernández-Meneses, M.; Puerta-Alcalde, P.; Chumbita, M.; Garcia-Pouton, N.; Linares, L.; Rico, V.; Cardozo, C.; Martínez, J.A.; et al. Personalized Therapy Approach for Hospitalized Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 74, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Carvalho, A.; Van De Veerdonk, F.L.; Jenks, J.D.; Koehler, P.; Krause, R.; Cornely, O.A.; Perlin, D.S.; Lass-Flörl, C.; Hoenigl, M. COVID-19 Associated Pulmonary Aspergillosis (CAPA)—From Immunology to Treatment. J. Fungi 2020, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- White, P.L.; Dhillon, R.; Cordey, A.; Hughes, H.; Faggian, F.; Soni, S.; Pandey, M.; Whitaker, H.; May, A.; Morgan, M.; et al. A National Strategy to Diagnose Coronavirus Disease 2019–Associated Invasive Fungal Disease in the Intensive Care Unit. Clin. Infect. Dis. 2020, 73, e1634–e1644. [Google Scholar] [CrossRef] [PubMed]
- Al-Hatmi, A.M.; Mohsin, J.; Al-Huraizi, A.; Khamis, F. COVID-19 associated invasive candidiasis. J. Infect. 2021, 82, e45–e46. [Google Scholar] [CrossRef]
- Antinori, S.; Galimberti, L.; Milazzo, L.; Ridolfo, A.L. Bacterial and Fungal Infections among Patients with SARS-CoV-2 Pneumonia. Le Infez. Med. 2020, 28, 29–36. [Google Scholar]
- Chowdhary, A.; Tarai, B.; Singh, A.; Sharma, A. Multidrug-Resistant Candida auris Infections in Critically Ill Coronavirus Disease Patients, India, April–July 2020. Emerg. Infect. Dis. 2020, 26, 2694–2696. [Google Scholar] [CrossRef]
- Mastrangelo, A.; Germinario, B.N.; Ferrante, M.; Frangi, C.; Voti, R.L.; Muccini, C.; Ripa, M.; Canetti, D.; Castiglioni, B.; Oltolini, C.; et al. Candidemia in Coronavirus Disease 2019 (COVID-19) Patients: Incidence and Characteristics in a Prospective Cohort Compared With Historical Non–COVID-19 Controls. Clin. Infect. Dis. 2020, 73, e2838–e2839. [Google Scholar] [CrossRef]
- Heard, K.L.; Hughes, S.; Mughal, N.; Moore, L. COVID-19 and fungal superinfection. Lancet Microbe 2020, 1, e107. [Google Scholar] [CrossRef]
- Moser, D.; Biere, K.; Han, B.; Hoerl, M.; Schelling, G.; Choukér, A.; Woehrle, T. COVID-19 Impairs Immune Response to Candida albicans. Front. Immunol. 2021, 12, 640644. [Google Scholar] [CrossRef]
- Singh, R.; Zogg, H.; Wei, L.; Bartlett, A.; Ghoshal, U.C.; Rajender, S.; Ro, S. Gut Microbial Dysbiosis in the Pathogenesis of Gastrointestinal Dysmotility and Metabolic Disorders. J. Neurogastroenterol. Motil. 2021, 27, 19–34. [Google Scholar] [CrossRef]
- Sharma, M.; Kaushal, K.; Rawat, S.S.; Muraleedharan, M.; Chhabra, S.; Verma, N.; Mittal, A.; Bahl, A.; Khullar, M.; Ramavat, A.; et al. The Cellular Stress Response Interactome and Extracellular Matirx Cross-Talk during Fibrosis: Stressed Extra-Matrix Affair. In Extracellular Matrix—Developments and Therapeutics; Madhurapantula, R.S., Orgel, J., Loewy, Z., Blumenberg, M., Eds.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Beatson, R.; Graham, R.; Freile, F.G.; Cozzetto, D.; Kannambath, S.; Pfeifer, E.; Woodman, N.; Owen, J.; Nuamah, R.; Mandel, U.; et al. Cancer-associated hypersialylated MUC1 drives the differentiation of human monocytes into macrophages with a pathogenic phenotype. Commun. Biol. 2020, 3, 644. [Google Scholar] [CrossRef]
- Petiz, L.L.; Glaser, T.; Scharfstein, J.; Ratajczak, M.Z.; Ulrich, H. P2Y14 Receptor as a Target for Neutrophilia Attenuation in Severe COVID-19 Cases: From Hematopoietic Stem Cell Recruitment and Chemotaxis to Thrombo-inflammation. Stem Cell Rev. Rep. 2021, 17, 241–252. [Google Scholar] [CrossRef]
- Chatterjee, M.; Huang, L.Z.; Wang, C.; Mykytyn, A.Z.; Westendorp, B.; Wubbolts, R.W.; Bosch, B.-J.; Haagmans, B.L.; van Putten, J.P.; Strijbis, K. The Glycosylated Extracellular Domain of MUC1 Protects against SARS-CoV-2 Infection at the Respiratory Surface. bioRxiv 2021. [Google Scholar] [CrossRef]
- Schepler, H.; Wang, X.; Neufurth, M.; Wang, S.; Schröder, H.C.; Müller, W.E.G. The therapeutic potential of inorganic polyphosphate: A versatile physiological polymer to control coronavirus disease (COVID-19). Theranostics 2021, 11, 6193–6213. [Google Scholar] [CrossRef]
- Karcz, T.P.; Whitehead, G.S.; Nakano, K.; Nakano, H.; Grimm, S.A.; Williams, J.G.; Deterding, L.J.; Jacobson, K.A.; Cook, D.N. UDP-glucose and P2Y14 receptor amplify allergen-induced airway eosinophilia. J. Clin. Investig. 2021, 131, e140709. [Google Scholar] [CrossRef]
- Jain, S.; Pydi, S.P.; Jung, Y.-H.; Scortichini, M.; Kesner, E.L.; Karcz, T.P.; Cook, D.N.; Gavrilova, O.; Wess, J.; Jacobson, K.A. Adipocyte P2Y14 receptors play a key role in regulating whole-body glucose and lipid homeostasis. JCI Insight 2021, 6, e146577. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, K.; Ma, L.; Qian, Z.; Tian, X.; Miao, Y.; Niu, Y.; Xu, X.; Guo, S.; Yang, Y.; et al. Endogenous glutamate determines ferroptosis sensitivity via ADCY10-dependent YAP suppression in lung adenocarcinoma. Theranostics 2021, 11, 5650–5674. [Google Scholar] [CrossRef]
- Taneri, P.E.; Gómez-Ochoa, S.A.; Llanaj, E.; Raguindin, P.F.; Rojas, L.Z.; Roa-Díaz, Z.M.; Salvador, D.; Groothof, D.; Minder, B.; Kopp-Heim, D. Anemia and iron metabolism in COVID-19: A systematic review and meta-analysis. Eur. J. Epidemiol. 2020, 35, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Sonnweber, T.; Boehm, A.; Sahanic, S.; Pizzini, A.; Aichner, M.; Sonnweber, B.; Kurz, K.; Koppelstätter, S.; Haschka, D.; Petzer, V.; et al. Persisting alterations of iron homeostasis in COVID-19 are associated with non-resolving lung pathologies and poor patients’ performance: A prospective observational cohort study. Respir. Res. 2020, 21, 276. [Google Scholar] [CrossRef] [PubMed]
- Carota, G.; Ronsisvalle, S.; Panarello, F.; Tibullo, D.; Nicolosi, A.; Volti, G.L. Role of Iron Chelation and Protease Inhibition of Natural Products on COVID-19 Infection. J. Clin. Med. 2021, 10, 2306. [Google Scholar] [CrossRef]
- Maiti, B.K. Heme/Hemeoxygenase-1 System Is a Potential Therapeutic Intervention for COVID-19 Patients with Severe Complications. ACS Pharmacol. Transl. Sci. 2020, 3, 1032–1034. [Google Scholar] [CrossRef]
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef]
- Combes, A.J.; Courau, T.; Kuhn, N.F.; Hu, K.H.; Ray, A.; Chen, W.S.; Cleary, S.J.; Chew, N.W.; Kushnoor, D.; Reeder, G.C.; et al. Global Absence and Targeting of Protective Immune States in Severe COVID-19. Nature 2021, 591, 124–130. [Google Scholar] [CrossRef]
- Lin, Z.; Long, F.; Yang, Y.; Chen, X.; Xu, L.; Yang, M. Serum ferritin as an independent risk factor for severity in COVID-19 patients. J. Infect. 2020, 81, 647–679. [Google Scholar] [CrossRef]
- Kusnadi, A.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Meckiff, B.J.; Simon, H.; Pelosi, E.; Seumois, G.; Ay, F.; Vijayanand, P.; et al. Severely ill patients with COVID-19 display impaired exhaustion features in SARS-CoV-2–reactive CD8 + T cells. Sci. Immunol. 2021, 6, eabe4782. [Google Scholar] [CrossRef]
- Huang, C.; Huang, L.; Wang, Y.; Li, X.; Ren, L.; Gu, X.; Kang, L.; Guo, L.; Liu, M.; Zhou, X.; et al. 6-month consequences of COVID-19 in patients discharged from hospital: A cohort study. Lancet 2021, 397, 220–232. [Google Scholar] [CrossRef]
- Carvalho, T.; Krammer, F.; Iwasaki, A. The first 12 months of COVID-19: A timeline of immunological insights. Nat. Rev. Immunol. 2021, 21, 245–256. [Google Scholar] [CrossRef]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Goel, R.R.; Apostolidis, S.A.; Painter, M.M.; Mathew, D.; Pattekar, A.; Kuthuru, O.; Gouma, S.; Hicks, P.; Meng, W.; Rosenfeld, A.M.; et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals after mRNA vaccination. Sci. Immunol. 2021, 6, eabi6950. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef] [Green Version]
- Teijaro, J.R.; Farber, D.L. COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef]
- Male, V. Are COVID-19 vaccines safe in pregnancy? Nat. Rev. Immunol. 2021, 21, 200–201. [Google Scholar] [CrossRef]
- Deinhardt-Emmer, S.; Wittschieber, D.; Sanft, J.; Kleemann, S.; Elschner, S.; Haupt, K.F.; Vau, V.; Häring, C.; Rödel, J.; Henke, A.; et al. Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage. eLife 2021, 10, e60361. [Google Scholar] [CrossRef]
- Smeda, M.; Chlopicki, S. Endothelial barrier integrity in COVID-19-dependent hyperinflammation: Does the protective facet of platelet function matter? Cardiovasc. Res. 2020, 116, e118–e121. [Google Scholar] [CrossRef]
- Shah, M.; Sachdeva, M.; Dodiuk-Gad, R.P. COVID-19 and racial disparities. J. Am. Acad. Dermatol. 2020, 83, e35. [Google Scholar] [CrossRef]
- López-Reyes, A.; Martinez-Armenta, C.; Espinosa-Velázquez, R.; Vázquez-Cárdenas, P.; Cruz-Ramos, M.; Palacios-Gonzalez, B.; Gomez-Quiroz, L.E.; Martínez-Nava, G.A. NLRP3 Inflammasome: The Stormy Link Between Obesity and COVID-19. Front. Immunol. 2020, 11, 570251. [Google Scholar] [CrossRef]
- Gedefaw, L.; Ullah, S.; Leung, P.; Cai, Y.; Yip, S.-P.; Huang, C.-L. Inflammasome Activation-Induced Hypercoagulopathy: Impact on Cardiovascular Dysfunction Triggered in COVID-19 Patients. Cells 2021, 10, 916. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef]
- Hoagland, D.A.; Møller, R.; Uhl, S.A.; Oishi, K.; Frere, J.; Golynker, I.; Horiuchi, S.; Panis, M.; Blanco-Melo, D.; Sachs, D.; et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity 2021, 54, 557–570.e5. [Google Scholar] [CrossRef]
- Yan, L.; Yang, Y.; Li, M.; Zhang, Y.; Zheng, L.; Ge, J.; Huang, Y.C.; Liu, Z.; Wang, T.; Gao, S.; et al. Coupling of N7-methyltransferase and 3′-5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell 2021, 184, 3474–3485.e11. [Google Scholar] [CrossRef]
- Balkhi, M.Y. Mechanistic understanding of innate and adaptive immune responses in SARS-CoV-2 infection. Mol. Immunol. 2021, 135, 268–275. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, Y.; Zhao, X.; Shao, L.; Liu, G.; Sun, C.; Xu, R.; Zhang, Z. ACSL4 exacerbates ischemic stroke by promoting ferroptosis-induced brain injury and neuroinflammation. Brain Behav. Immun. 2021, 93, 312–321. [Google Scholar] [CrossRef]
- Gómez-Henao, W.; Tenorio, E.P.; Sanchez, F.R.C.; Mendoza, M.C.; Ledezma, R.L.; Zenteno, E. Relevance of glycans in the interaction between T lymphocyte and the antigen presenting cell. Int. Rev. Immunol. 2020, 40, 274–288. [Google Scholar] [CrossRef]
- DeRosa, A.; Leftin, A. The Iron Curtain: Macrophages at the Interface of Systemic and Microenvironmental Iron Metabolism and Immune Response in Cancer. Front. Immunol. 2021, 12, 614294. [Google Scholar] [CrossRef]
- Winn, B.J. Is there a role for insulin-like growth factor inhibition in the treatment of COVID-19-related adult respiratory distress syndrome? Med. Hypotheses 2020, 144, 110167. [Google Scholar] [CrossRef]
- Cronin, S.J.F.; Woolf, C.J.; Weiss, G.; Penninger, J.M. The Role of Iron Regulation in Immunometabolism and Immune-Related Disease. Front. Mol. Biosci. 2019, 6, 116. [Google Scholar] [CrossRef] [Green Version]
- Weiss-Sadan, T.; Maimoun, D.; Oelschlagel, D.; Kaschani, F.; Misiak, D.; Gaikwad, H.; Ben-Nun, Y.; Merquiol, E.; Anaki, A.; Tsvirkun, D.; et al. Cathepsins Drive Anti-Inflammatory Activity by Regulating Autophagy and Mitochondrial Dynamics in Macrophage Foam Cells. Cell. Physiol. Biochem. 2019, 53, 550–572. [Google Scholar] [CrossRef] [Green Version]
- Nairz, M.; Weiss, G. Iron in infection and immunity. Mol. Asp. Med. 2020, 75, 100864. [Google Scholar] [CrossRef]
- Katsarou, A.; Pantopoulos, K. Basics and principles of cellular and systemic iron homeostasis. Mol. Asp. Med. 2020, 75, 100866. [Google Scholar] [CrossRef]
- Mochochoko, B.M.; Ezeokoli, O.T.; Sebolai, O.; Albertyn, J.; Pohl, C.H. Role of the high-affinity reductive iron acquisition pathway of Candida albicans in prostaglandin E2 production, virulence, and interaction with Pseudomonas aeruginosa. Med. Mycol. 2021, 59, 869–881. [Google Scholar] [CrossRef]
- Perea-García, A.; Borderia, D.A.; Vera-Sirera, F.; Pérez-Amador, M.A.; Puig, S.; Peñarrubia, L. Deregulated High Affinity Copper Transport Alters Iron Homeostasis in Arabidopsis. Front. Plant Sci. 2020, 11, 1106. [Google Scholar] [CrossRef]
- Stanford, F.; Matthies, N.; Cseresnyés, Z.; Figge, M.; Hassan, M.; Voigt, K. Expression Patterns in Reductive Iron Assimilation and Functional Consequences during Phagocytosis of Lichtheimia corymbifera, an Emerging Cause of Mucormycosis. J. Fungi 2021, 7, 272. [Google Scholar] [CrossRef]
- Jung, J.H.; Rha, M.-S.; Sa, M.; Choi, H.K.; Jeon, J.H.; Seok, H.; Park, D.W.; Park, S.-H.; Jeong, H.W.; Choi, W.S.; et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat. Commun. 2021, 12, 4043. [Google Scholar] [CrossRef]
- Kumar, H.M.; Sharma, P.; Rudramurthy, S.M.; Sehgal, I.S.; Prasad, K.T.; Pannu, A.K.; Das, R.; Panda, N.K.; Sharma, N.; Chakrabarti, A. Serum iron indices in COVID-19-associated mucormycosis: A case–control study. Mycoses 2021, 65, 120–127. [Google Scholar] [CrossRef]
- Galaris, D.; Barbouti, A.; Pantopoulos, K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim. Biophys. Acta 2019, 1866, 118535. [Google Scholar] [CrossRef]
- Maio, N.; Rouault, T.A. Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis. Trends Biochem. Sci. 2020, 45, 411–426. [Google Scholar] [CrossRef]
- Dickson-Murray, E.; Nedara, K.; Modjtahedi, N.; Tokatlidis, K. The Mia40/CHCHD4 Oxidative Folding System: Redox Regulation and Signaling in the Mitochondrial Intermembrane Space. Antioxidants 2021, 10, 592. [Google Scholar] [CrossRef]
- Berndt, N.; Kolbe, E.; Gajowski, R.; Eckstein, J.; Ott, F.; Meierhofer, D.; Holzhütter, H.; Matz-Soja, M. Functional Consequences of Metabolic Zonation in Murine Livers: Insights for an Old Story. Hepatology 2021, 73, 795–810. [Google Scholar] [CrossRef] [Green Version]
- Talib, E.A.; Outten, C.E. Iron-sulfur cluster biogenesis, trafficking, and signaling: Roles for CGFS glutaredoxins and BolA proteins. Biochim. Biophys. Acta 2021, 1868, 118847. [Google Scholar] [CrossRef]
- Braymer, J.J.; Freibert, S.A.; Rakwalska-Bange, M.; Lill, R. Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118863. [Google Scholar] [CrossRef]
- Daniel, T.; Faruq, H.M.; Magdalena, J.L.; Manuela, G.; Horst, L.C. Role of GSH and Iron-Sulfur Glutaredoxins in Iron Metabolism—Review. Molecules 2020, 25, 3860. [Google Scholar] [CrossRef]
- Vogt, A.-C.; Arsiwala, T.; Mohsen, M.; Vogel, M.; Manolova, V.; Bachmann, M. On Iron Metabolism and Its Regulation. Int. J. Mol. Sci. 2021, 22, 4591. [Google Scholar] [CrossRef]
- Liu, T.; Pei, K.; Wang, Z.; Wang, Z.-L. Pivotal effects of external Fe2+ on remediation of arsenite by zero-valent iron/persulfate: Efficiencies and mechanism. Environ. Res. 2020, 189, 109922. [Google Scholar] [CrossRef]
- Han, V.X.; Jones, H.F.; Patel, S.; Mohammad, S.S.; Hofer, M.J.; Alshammery, S.; Maple-Brown, E.; Gold, W.; Brilot, F.; Dale, R.C. Emerging evidence of Toll-like receptors as a putative pathway linking maternal inflammation and neurodevelopmental disorders in human offspring: A systematic review. Brain, Behav. Immun. 2021, 99, 91–105. [Google Scholar] [CrossRef]
- Elgendy, S.M.; Alyammahi, S.K.; Alhamad, D.W.; Abdin, S.M.; Omar, H.A. Ferroptosis: An emerging approach for targeting cancer stem cells and drug resistance. Crit. Rev. Oncol. 2020, 155, 103095. [Google Scholar] [CrossRef]
- Anthonymuthu, T.S.; Tyurina, Y.Y.; Sun, W.-Y.; Mikulska-Ruminska, K.; Shrivastava, I.H.; Tyurin, V.A.; Cinemre, F.B.; Dar, H.H.; VanDemark, A.P.; Holman, T.R.; et al. Resolving the paradox of ferroptotic cell death: Ferrostatin-1 binds to 15LOX/PEBP1 complex, suppresses generation of peroxidized ETE-PE, and protects against ferroptosis. Redox Biol. 2020, 38, 101744. [Google Scholar] [CrossRef]
- Tang, D.; Kroemer, G. Ferroptosis. Curr. Biol. 2020, 30, R1292–R1297. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Jiang, X.; Gu, W. Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cell Biol. 2020, 30, 478–490. [Google Scholar] [CrossRef]
- Tang, R.; Xu, J.; Zhang, B.; Liu, J.; Liang, C.; Hua, J.; Meng, Q.; Yu, X.; Shi, S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J. Hematol. Oncol. 2020, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Elbadawi, M.; Efferth, T. Multiple cell death modalities and their key features (Review). World Acad. Sci. J. 2020, 2, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Dai, C.; Chen, X.; Li, J.; Comish, P.; Kang, R.; Tang, D. Transcription factors in ferroptotic cell death. Cancer Gene Ther. 2020, 27, 645–656. [Google Scholar] [CrossRef]
- Song, X.; Liu, J.; Kuang, F.; Chen, X.; Zeh, H.J.; Kang, R.; Kroemer, G.; Xie, Y.; Tang, D. PDK4 dictates metabolic resistance to ferroptosis by suppressing pyruvate oxidation and fatty acid synthesis. Cell Rep. 2021, 34, 108767. [Google Scholar] [CrossRef]
- Zeitler, L.; Fiore, A.; Meyer, C.; Russier, M.; Zanella, G.; Suppmann, S.; Gargaro, M.; Sidhu, S.S.; Seshagiri, S.; Ohnmacht, C.; et al. Anti-ferroptotic mechanism of IL4i1-mediated amino acid metabolism. eLife 2021, 10, e64806. [Google Scholar] [CrossRef]
- Lopes-Coelho, F.; Martins, F.; Hipólito, A.; Mendes, C.; Sequeira, C.O.; Pires, R.F.; Almeida, A.M.; Bonifácio, V.D.B.; Pereira, S.A.; Serpa, J. The Activation of Endothelial Cells Relies on a Ferroptosis-Like Mechanism: Novel Perspectives in Management of Angiogenesis and Cancer Therapy. Front. Oncol. 2021, 11, 656229. [Google Scholar] [CrossRef]
- Chen, L.; Hambright, W.S.; Na, R.; Ran, Q. Ablation of the Ferroptosis Inhibitor Glutathione Peroxidase 4 in Neurons Results in Rapid Motor Neuron Degeneration and Paralysis. J. Biol. Chem. 2015, 290, 28097–28106. [Google Scholar] [CrossRef] [Green Version]
- Kapralov, A.A.; Yang, Q.; Dar, H.H.; Tyurina, Y.Y.; Anthonymuthu, T.S.; Kim, R.; St Croix, C.M.; Mikulska-Ruminska, K.; Liu, B.; Shrivastava, I.H.; et al. Redox lipid reprogramming commands susceptibility of macrophages and microglia to ferroptotic death. Nat. Chem. Biol. 2020, 16, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Tonnus, W.; Meyer, C.; Paliege, A.; Belavgeni, A.; Von Mässenhausen, A.; Bornstein, S.R.; Hugo, C.; Becker, J.U.; Linkermann, A. The pathological features of regulated necrosis. J. Pathol. 2019, 247, 697–707. [Google Scholar] [CrossRef]
- Bayır, H.; Anthonymuthu, T.S.; Tyurina, Y.; Patel, S.J.; Amoscato, A.A.; Lamade, A.M.; Yang, Q.; Vladimirov, G.K.; Philpott, C.C.; Kagan, V.E. Achieving Life through Death: Redox Biology of Lipid Peroxidation in Ferroptosis. Cell Chem. Biol. 2020, 27, 387–408. [Google Scholar] [CrossRef] [PubMed]
- Mleczko-Sanecka, K.; Silvestri, L. Cell-type-specific insights into iron regulatory processes. Am. J. Hematol. 2021, 96, 110–127. [Google Scholar] [CrossRef] [PubMed]
- Camaschella, C.; Nai, A.; Silvestri, L. Iron metabolism and iron disorders revisited in the hepcidin era. Haematologica 2020, 105, 260–272. [Google Scholar] [CrossRef] [Green Version]
- Stockwell, B.R.; Angeli, J.P.F.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Dar, M.A.; Hölscher, C. Arginase-1 Is Responsible for IL-13-Mediated Susceptibility to Trypanosoma cruzi Infection. Front. Immunol. 2018, 9, 2790. [Google Scholar] [CrossRef]
- Amaral, E.P.; Costa, D.L.; Namasivayam, S.; Riteau, N.; Kamenyeva, O.; Mittereder, L.; Mayer-Barber, K.D.; Andrade, B.B.; Sher, A. A major role for ferroptosis in Mycobacterium tuberculosis–induced cell death and tissue necrosis. J. Exp. Med. 2019, 216, 556–570. [Google Scholar] [CrossRef]
- Horwath, M.C.; Bell-Horwath, T.R.; Lescano, V.; Krishnan, K.; Merino, E.J.; Deepe, G.S. Antifungal Activity of the Lipophilic Antioxidant Ferrostatin-1. ChemBioChem 2017, 18, 2069–2078. [Google Scholar] [CrossRef]
- Stoyanovsky, D.; Tyurina, Y.; Shrivastava, I.; Bahar, I.; Tyurin, V.; Protchenko, O.; Jadhav, S.; Bolevich, S.; Kozlov, A.; Vladimirov, Y.; et al. Iron catalysis of lipid peroxidation in ferroptosis: Regulated enzymatic or random free radical reaction? Free Radic. Biol. Med. 2019, 133, 153–161. [Google Scholar] [CrossRef]
- Patel, S.J.; Frey, A.G.; Palenchar, D.J.; Achar, S.; Bullough, K.Z.; Vashisht, A.; Wohlschlegel, J.A.; Philpott, C.C. A PCBP1–BolA2 chaperone complex delivers iron for cytosolic [2Fe–2S] cluster assembly. Nat. Chem. Biol. 2019, 15, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Philpott, C.C.; Jadhav, S. The ins and outs of iron: Escorting iron through the mammalian cytosol. Free Radic. Biol. Med. 2019, 133, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.R.; Eid, R.; Miller, K.A.; Boucher, E.; Mandato, C.A.; Greenwood, M.T. Intracellular second messengers mediate stress inducible hormesis and Programmed Cell Death: A review. Biochim. Biophys. Acta 2019, 1866, 773–792. [Google Scholar] [CrossRef] [PubMed]
- Pierzynowska, K.; Rintz, E.; Gaffke, L.; Węgrzyn, G. Ferroptosis and Its Modulation by Autophagy in Light of the Pathogenesis of Lysosomal Storage Diseases. Cells 2021, 10, 365. [Google Scholar] [CrossRef]
- Ajoolabady, A.; Aslkhodapasandhokmabad, H.; Libby, P.; Tuomilehto, J.; Lip, G.Y.; Penninger, J.M.; Richardson, D.R.; Tang, D.; Zhou, H.; Wang, S.; et al. Ferritinophagy and ferroptosis in the management of metabolic diseases. Trends Endocrinol. Metab. 2021, 32, 444–462. [Google Scholar] [CrossRef]
- Willenborg, S.; Sanin, D.E.; Jais, A.; Ding, X.; Ulas, T.; Nüchel, J.; Popović, M.; MacVicar, T.; Langer, T.; Schultze, J.L.; et al. Mitochondrial metabolism coordinates stage-specific repair processes in macrophages during wound healing. Cell Metab. 2021, 33, 2398–2414.e9. [Google Scholar] [CrossRef]
- Cougnon, M.; Carcy, R.; Melis, N.; Rubera, I.; Duranton, C.; Dumas, K.; Tanti, J.-F.; Pons, C.; Soubeiran, N.; Shkreli, M.; et al. Inhibition of eIF5A hypusination reprogrammes metabolism and glucose handling in mouse kidney. Cell Death Dis. 2021, 12, 283. [Google Scholar] [CrossRef]
- Jeelani, G.; Nozaki, T. Eukaryotic translation initiation factor 5A and its posttranslational modifications play an important role in proliferation and potentially in differentiation of the human enteric protozoan parasite Entamoeba histolytica. PLoS Pathog. 2021, 17, e1008909. [Google Scholar] [CrossRef]
- Nakamura, A.; Kurihara, S.; Takahashi, D.; Ohashi, W.; Nakamura, Y.; Kimura, S.; Onuki, M.; Kume, A.; Sasazawa, Y.; Furusawa, Y.; et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat. Commun. 2021, 12, 2105. [Google Scholar] [CrossRef]
- Liang, Y.; Piao, C.; Beuschel, C.B.; Toppe, D.; Kollipara, L.; Bogdanow, B.; Maglione, M.; Lützkendorf, J.; See, J.C.K.; Huang, S.; et al. eIF5A hypusination, boosted by dietary spermidine, protects from premature brain aging and mitochondrial dysfunction. Cell Rep. 2021, 35, 108941. [Google Scholar] [CrossRef]
- Medina, C.B.; Mehrotra, P.; Arandjelovic, S.; Perry, J.S.A.; Guo, Y.; Morioka, S.; Barron, B.; Walk, S.F.; Ghesquière, B.; Krupnick, A.S.; et al. Metabolites released from apoptotic cells act as tissue messengers. Nature 2020, 580, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Eisenberg, T.; Pietrocola, F.; Kroemer, G. Spermidine in health and disease. Science 2018, 359, eaan2788. [Google Scholar] [CrossRef] [Green Version]
- Ou, Y.; Wang, S.-J.; Li, D.; Chu, B.; Gu, W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. USA 2016, 113, E6806–E6812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Zhang, W.; Chen, H.; Zeng, J. Targeting Polyamine Metabolism for Control of Human Viral Diseases. Infect. Drug Resist. 2020, 13, 4335–4346. [Google Scholar] [CrossRef]
- Yoshinaga, M.; Takeuchi, O. RNA binding proteins in the control of autoimmune diseases. Immunol. Med. 2019, 42, 53–64. [Google Scholar] [CrossRef] [Green Version]
- Ansa-Addo, E.A.; Huang, H.-C.; Riesenberg, B.; Iamsawat, S.; Borucki, D.; Nelson, M.H.; Nam, J.H.; Chung, D.; Paulos, C.M.; Liu, B.; et al. RNA binding protein PCBP1 is an intracellular immune checkpoint for shaping T cell responses in cancer immunity. Sci. Adv. 2020, 6, eaaz3865. [Google Scholar] [CrossRef] [PubMed]
- Cornelius, V.A.; Yacoub, A.; Kelaini, S.; Margariti, A. Diabetic endotheliopathy: RNA-binding proteins as new therapeutic targets. Int. J. Biochem. Cell Biol. 2021, 131, 105907. [Google Scholar] [CrossRef]
- Moss, N.D.; Sussel, L. mRNA Processing: An Emerging Frontier in the Regulation of Pancreatic β Cell Function. Front. Genet. 2020, 11, 983. [Google Scholar] [CrossRef]
- Cao, J.Y.; Dixon, S.J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. 2016, 73, 2195–2209. [Google Scholar] [CrossRef] [Green Version]
- Nie, A.; Sun, B.; Fu, Z.; Yu, D. Roles of aminoacyl-tRNA synthetases in immune regulation and immune diseases. Cell Death Dis. 2019, 10, 901. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Sun, B.; Huang, S.; Jia, W.; Yu, D. The tRNA-associated dysregulation in diabetes mellitus. Metabolism 2019, 94, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, M.N.; Green, J.A.; Cnop, M.; Igoillo-Esteve, M. tRNA Biology in the Pathogenesis of Diabetes: Role of Genetic and Environmental Factors. Int. J. Mol. Sci. 2021, 22, 496. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Chatterjee, S.; Das, A.; Panda, N.; Shivaprakash, M.; Kaur, A.; Varma, S.; Singhi, S.; Bhansali, A.; Sakhuja, V. Invasive zygomycosis in India: Experience in a tertiary care hospital. Postgrad. Med. J. 2009, 85, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Targeted Environmental Investigation Checklist for Outbreaks of Invasive Infections Caused by Environmental Fungi (e.g., Aspergillus, Mucormycetes). Available online: https://www.cdc.gov/fungal/pdf/targeted-environmental-investigation-checklist-508.pdf (accessed on 12 January 2022).
- Bassetti, M.; Azoulay, E.; Kullberg, B.-J.; Ruhnke, M.; Shoham, S.; Vazquez, J.; Giacobbe, D.R.; Calandra, T. EORTC/MSGERC Definitions of Invasive Fungal Diseases: Summary of Activities of the Intensive Care Unit Working Group. Clin. Infect. Dis. 2021, 72, S121–S127. [Google Scholar] [CrossRef]
- Honavar, S. Code Mucor: Guidelines for the Diagnosis, Staging and Management of Rhino-Orbito-Cerebral Mucormycosis in the Setting of COVID-19. Indian J. Ophthalmol. 2021, 69, 1361–1365. [Google Scholar] [CrossRef]
- Dolatabadi, S.; Walther, G.; Ende, A.H.G.G.V.D.; de Hoog, G.S. Diversity and delimitation of Rhizopus microsporus. Fungal Divers. 2014, 64, 145–163. [Google Scholar] [CrossRef]
- Potenza, L.; Vallerini, D.; Barozzi, P.; Riva, G.; Gilioli, A.; Forghieri, F.; Candoni, A.; Cesaro, S.; Quadrelli, C.; Maertens, J.; et al. Mucorales-Specific T Cells in Patients with Hematologic Malignancies. PLoS ONE 2016, 11, e0149108. [Google Scholar] [CrossRef] [Green Version]
- Afzali, B.; Noris, M.; Lambrecht, B.N.; Kemper, C. The state of complement in COVID-19. Nat. Rev. Immunol. 2021, 22, 77–84. [Google Scholar] [CrossRef]
- Lee, S.-J.; Depoortere, I.; Hatt, H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2018, 18, 116–138. [Google Scholar] [CrossRef]
- Tong, T.; Wang, Y.; Kang, S.-G.; Huang, K. Ectopic Odorant Receptor Responding to Flavor Compounds: Versatile Roles in Health and Disease. Pharmaceutics 2021, 13, 1314. [Google Scholar] [CrossRef]
- De Virgiliis, F.; Di Giovanni, S. Lung innervation in the eye of a cytokine storm: Neuroimmune interactions and COVID-19. Nat. Rev. Neurol. 2020, 16, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Conde, S.V.; Sacramento, J.F.; Martins, F.O. Immunity and the carotid body: Implications for metabolic diseases. Bioelectron. Med. 2020, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Iturriaga, R.; Del Rio, R.; Alcayaga, J. Carotid Body Inflammation: Role in Hypoxia and in the Anti-inflammatory Reflex. Physiology 2021. [Google Scholar] [CrossRef] [PubMed]
- Dalangin, R.; Kim, A.; Campbell, R.E. The Role of Amino Acids in Neurotransmission and Fluorescent Tools for Their Detection. Int. J. Mol. Sci. 2020, 21, 6197. [Google Scholar] [CrossRef] [PubMed]
- Orecchioni, M.; Kobiyama, K.; Winkels, H.; Ghosheh, Y.; McArdle, S.; Mikulski, Z.; Kiosses, W.B.; Fan, Z.; Wen, L.; Jung, Y.; et al. Olfactory receptor 2 in vascular macrophages drives atherosclerosis by NLRP3-dependent IL-1 production. Science 2022, 375, 214–221. [Google Scholar] [CrossRef]
- SMART—Servier Medical ART. Available online: https://smart.servier.com/ (accessed on 7 January 2022).
- Free COVID-19 (SARS-CoV-2) Illustrations. Available online: https://innovativegenomics.org/free-covid-19-illustrations/ (accessed on 7 January 2022).



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, M.; Vanam, H.P.; Panda, N.K.; Patro, S.K.; Arora, R.; Bhadada, S.K.; Rudramurthy, S.M.; Singh, M.P.; Koppula, P.R. Deciphering the Neurosensory Olfactory Pathway and Associated Neo-Immunometabolic Vulnerabilities Implicated in COVID-Associated Mucormycosis (CAM) and COVID-19 in a Diabetes Backdrop—A Novel Perspective. Diabetology 2022, 3, 193-235. https://doi.org/10.3390/diabetology3010013
Sharma M, Vanam HP, Panda NK, Patro SK, Arora R, Bhadada SK, Rudramurthy SM, Singh MP, Koppula PR. Deciphering the Neurosensory Olfactory Pathway and Associated Neo-Immunometabolic Vulnerabilities Implicated in COVID-Associated Mucormycosis (CAM) and COVID-19 in a Diabetes Backdrop—A Novel Perspective. Diabetology. 2022; 3(1):193-235. https://doi.org/10.3390/diabetology3010013
Chicago/Turabian StyleSharma, Maryada, Hari Pankaj Vanam, Naresh K. Panda, Sourabha K. Patro, Rhythm Arora, Sanjay K. Bhadada, Shivaprakash M. Rudramurthy, Mini P. Singh, and Purushotham Reddy Koppula. 2022. "Deciphering the Neurosensory Olfactory Pathway and Associated Neo-Immunometabolic Vulnerabilities Implicated in COVID-Associated Mucormycosis (CAM) and COVID-19 in a Diabetes Backdrop—A Novel Perspective" Diabetology 3, no. 1: 193-235. https://doi.org/10.3390/diabetology3010013
APA StyleSharma, M., Vanam, H. P., Panda, N. K., Patro, S. K., Arora, R., Bhadada, S. K., Rudramurthy, S. M., Singh, M. P., & Koppula, P. R. (2022). Deciphering the Neurosensory Olfactory Pathway and Associated Neo-Immunometabolic Vulnerabilities Implicated in COVID-Associated Mucormycosis (CAM) and COVID-19 in a Diabetes Backdrop—A Novel Perspective. Diabetology, 3(1), 193-235. https://doi.org/10.3390/diabetology3010013







