Abstract
The adaptable nature of the SARS-CoV-2 virus has led to the emergence of multiple viral variants of concern. This research builds upon a previous demonstration of sampling human hand odor to distinguish SARS-CoV-2 infection status in order to incorporate considerations of the disease variants. This study demonstrates the ability of human odor expression to be implemented as a non-invasive medium for the differentiation of SARS-CoV-2 variants. Volatile organic compounds (VOCs) were extracted from SARS-CoV-2-positive samples using solid phase microextraction (SPME) coupled with gas chromatography–mass spectrometry (GC–MS). Sparse partial least squares discriminant analysis (sPLS-DA) modeling revealed that supervised machine learning could be used to predict the variant identity of a sample using VOC expression alone. The class discrimination of Delta and Omicron BA.5 variant samples was performed with 95.2% (±0.4) accuracy. Omicron BA.2 and Omicron BA.5 variants were correctly classified with 78.5% (±0.8) accuracy. Lastly, Delta and Omicron BA.2 samples were assigned with 71.2% (±1.0) accuracy. This work builds upon the framework of non-invasive techniques producing diagnostics through the analysis of human odor expression, all in support of public health monitoring.
1. Introduction
The rapid screening and diagnosis of persons infected with the severe, acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus that causes the COVID-19 disease, has been a focus of the medical community since the start of the 2019 global pandemic. Due to the high demand for a diagnosis of viral infectivity, most existing diagnostic work has been solely aimed at determining the presence or absence of SARS-CoV-2 infection. Unfortunately, due to its pervasive nature, and thus its rapid replication rate, the SARS-CoV-2 virus has mutated multiple times since the start of the pandemic, with the most notable variants of concern afflicting North America (at the time of this work) being the Alpha, Beta, Delta, Omicron, and, more recently, the Omicron BA.5 variants [1]. This has required researchers to improve the diagnostic methods in order to adapt to the ever-evolving variants [2]. As defined by the Centers for Disease Control and Prevention (CDC), a variant is considered to be a “variant of concern” when there is clear evidence that the new mutation has led to a quicker transmission rate, higher mortality rate, increased diagnostic failures, or other similar traits of that nature [3]. Previously, various genomic sequencing technologies have been adapted for the detection and identification of variants for other viruses, such as hepatitis C [4], parvovirus B19 [5], and infectious bronchitis [6]. Similar sequencing technology has been used by the CDC for SARS-CoV-2 variant differentiation [7]. In addition to using genome sequencing, reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) has been advanced to distinguish between some of the virus variants [8,9]. However, advances in this area have been limited, and they require invasive sample collection. This has previously been circumvented in studies of COVID-19 diagnosis through the non-invasive sampling of volatile organic compounds (VOCs) from human breath [10,11] or body odor expression [12]. Secondary odor in human scent is commonly affected by metabolic and environmental causes, such as the menstrual cycle, diet, stress, and other factors [13]. Diseases, such as SARS-CoV-2, have been noted to have significant influences on human metabolism that cause a change in the expression of their secondary odor [14,15]. In 2022, Woollam et al. was able to combine multivariate data analysis with the chemical analysis of human breath to reliably distinguish between SARS-CoV-2-infected and healthy individuals [15]. Additionally, the authors have demonstrated a similar proof-of-concept utilizing human hand odor samples [16]. However, the researchers have not identified any literature relating to the separation of SARS-CoV-2 variants through human odor expression.
This is the second in a set of investigations that expands upon the potential use of human hand odor expression as a non-invasive diagnostic approach for SARS-CoV-2. The previous manuscript in this series established the viability of the use of human hand odor to predict SARS-CoV-2 infection status [16]. This particular work describes a novel approach for the differentiation of SARS-CoV-2 variants using supervised machine learning coupled to headspace- solid phase microextraction (HS-SPME) sampling and gas chromatography–mass spectrometry (GC–MS) analysis. The approach allows for the gaseous components from a heated hand sweat sample to be analyzed instrumentally for the identification of its chemical composition and interpreted for an indication of whether the SARS-CoV-2-positive sample is indicative of the Delta, Omicron BA.2, or Omicron BA.5 variants. It is crucial to distinguish between such variants, as this will provide surveillance programs with key information to guide our understanding of how the virus is spread, how quickly it mutates, and will ultimately inform the development of potential mitigation programs. This work looks to build upon the framework of non-invasive diagnostic techniques through the analysis of human odor expression as a means of supporting viral genomic surveillance and public health monitoring.
2. Materials and Methods
2.1. Human Hand Odor Collection
Human hand odor samples were collected from COVID-19-positive patients between June 2021 and September 2022. The samples were collected during three distinct collection periods that corresponded to periods of high infection rates for three different COVID-19 variants (Delta, Omicron BA.2, and Omicron BA.5), according to CDC data [1]. The variants were not confirmed via any genomic sequencing and, as such, are considered to be “suspected variants” for each sample. All samples were collected from patients in the Emergency Department at the Penn Presbyterian Medical Center (PPMC) by the Penn Acute Research Collaboration (PARC). Researchers at PARC supervised the self-donation of hand odor samples in accordance with the method approved by the University of Pennsylvania’s Institutional Review Board (IRB# 848819). The samples were analyzed by researchers at Florida International University in Miami, Florida, using HS-SPME- GC–MS.
2.2. Confirmation of SARS-CoV-2 Infection
In this study, 50 patients were tested for and diagnosed with SARS-CoV-2 infection using the Roche cobas® SARS-CoV-2 Duo Test for use on the cobas® 6800/8800 systems (Roche Diagnostics, Basel, Switzerland). The healthcare provider collected nasopharyngeal swab samples from all 56 patients for this PCR diagnostic test, which is the standard SARS-CoV-2 diagnostic method used by PPMC. Positive results were reported, per the FDA EUA instructions for use, and signified the presence of SARS-CoV-2 RNA [17]. Three (3) patients were tested using the Abbott BinaxNOW antigen test (Abbott Laboratories, Chicago, IL, USA), which consisted of a self-collected anterior nasal swab specimen. The results of this test indicated whether the nucleocapsid protein antigen of SARS-CoV-2 was identified [18]. Finally, the remaining three (3) patients were not tested by the participating researchers; while the results of their SARS-CoV-2 test were relayed to the research team, the sample specimen and test type remained unknown.
2.3. Sample Collection
The human hand odor samples were collected into pre-treated 10 mL headspace glass vials using sterile gauze pads of 2′′ by 2′′ and 8-ply density (Dukal Corporation, Syosset, NY) following vial cleaning and gauze pretreatment procedures as defined by Crespo-Cajigas [16]. For each patient, the sample gauze was placed inside of a 10 mL vial, which was contained inside an aluminum barrier bag. Both the vials and the bags were labeled with the following: (a) Sample #, (b) Date, (c) Sex at Birth, and (d) COVID-19: Positive.
2.4. Patient Demographics
The patients were asked to verbally disclose their age, race/ethnicity, sex at birth, symptomology, vaccination status, and chief health complaint (their primary reason for visiting the Emergency Department). The patient demographic information can be found in the Appendix A in Table A1. Table 1 notes the timeframe for three collection timespans observed by the study.

Table 1.
COVID-19-Positive Samples.
2.5. HS-SPME- GC–MS Method
The extraction of the VOCs from the samples started with heating the samples contained within sealed headspace vials at 50 °C in a digital heating bath (Thermo Fisher Scientific, Waltham, MA, USA). After 24 h, a SPME fiber with a 50/30 µm divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) coating was placed through the septum of the heated vial and exposed to the sample headspace at a height of 1 cm. The SPME fiber was exposed to the sample headspace for 15 h at 50 °C. Once the extraction was completed, the SPME fiber was placed directly into the heated GC inlet (Agilent 8890; Agilent Technologies, Santa Clara, CA, USA) for 5 min at 270 °C for the desorption of the analytes (2 cm height). A splitless injection method with a 1 mL/min column flow was implemented on an HP5-MS UI capillary column (15 m × 0.250 mm × 0.25 µm I.D.; Agilent Technologies). UHP helium was used as the carrier gas. The oven temperature parameters started at 40 °C (1.25 min hold), increased to 165 °C (5 °C/min rate), and concluded at 270 °C (30 °C/min rate). The total method runtime was 29.75 min. A mass spectrometer (MS) (Agilent 5977B MSD; Agilent Technologies, Santa Clara, CA, USA) with an electron impact ionization (EI) source and quadrupole mass analyzer was used with the following parameters: the MS source was maintained at 230 °C, the MS Quad at 150 °C, the transfer line at 280 °C, and the EI source at 70 eV. The samples collected between June 2021 and May 2022 were analyzed using a scan range of m/z 50–550. The samples analyzed between July 2022 and October 2022 were analyzed using a 45–400 m/z scan range.
2.6. Data Pre-Processing
The human hand odor samples were analyzed using the HS-SPME- GC–MS method outlined in Section 2.5. The analyzed samples were represented by the chemical characterization of the gaseous components of each sample. The data files resulting from the instrumental analysis were pre-processed with retention time alignment and peak matching across the dataset. The data files were background subtracted using the relevant sample blank (Delta samples) or batch method blank (Omicron BA.2 and Omicron BA.5 samples). The sample blanks consisted of unused collection materials that were packaged alongside the collected patient sample. They were submitted in reference to a specific collected patient sample (n = 1 per patient); whereas, the batch method blank consisted of unused sample materials that were pretreated and prepared by the researchers alongside the pretreated collection materials. The method blank samples were unused collection materials that were stored and transported in reference to all samples collected within a defined window of time (n = 3 blanks per batch). Compounds found to be present in the respective reference blank sample were removed from consideration in the analysis step.
This procedure was followed by log-10 transformation of total ion chromatogram peak areas of the aligned and matched peaks; furthermore, all values of 0 were set to 1 before applying the transformation. A total of 40 key features of interest were identified in a previous publication [16] for their role in predicting COVID-19 infection status using human hand odor. These 40 features of interest were monitored in this current work and interpreted for the variant-specific expression demonstrated by each COVID-19 subvariant.
2.7. Features of Interest
A total of 40 key features of interest were tracked using retention time alignment and peak matching across the dataset. Of these 40 features, the following 14 compounds were identified: (#5) Diacetone alcohol (3.109 min, CAS#123-42-2), (#6) Styrene (4.033 min, CAS#100-42-5), (#10) 2-Pentylfuran (6.577 min, CAS#3777-69-3), (#12) Phenylacetaldehyde (7.897 min, CAS#122-78-1), (#20) Undecane (9.552 min, CAS#1120-21-4), (#24) Methyl caprylate (10.366 min, CAS#111-11-5), (#26) trans-2-Nonenal (11.235 min, CAS#18829-56-6), (#27) 1-Nonanol (11.679 min, CAS#143-08-7), (#32) Decanal (12.561 min, CAS#112-31-2), (#33) 2-Phenoxyethanol (12.836 min, CAS#122-99-6), (#35) Dodecanal (17.798 min, CAS#112-54-9), (#37) 6,10-Dimethyl-5,9-undecadien-2-one-(E) (18.841 min, CAS#689-67-8), (#38) 1-Dodecanol (19.366 min, CAS#112-53-8), and (#39) Dodecanoic acid (21.668 min, CAS#143-07-7). The locations of all 40 features of interest are highlighted in Figure 1, which demonstrates overlayed chromatograms corresponding to the hand odor of one patient from each of the Delta, Omicron BA.2, and Omicron BA.5 datasets. Each feature of interest is not present in each sample, and, as such, is not present in each chromatogram in Figure 1. The average trend of these features of interest across each dataset is graphically represented in Figure A1. The absence, or reduced presence, of some of the features across the datasets are graphically displayed using the log-10 transformed average peak areas of the 40 features of interest, separated by each variant.
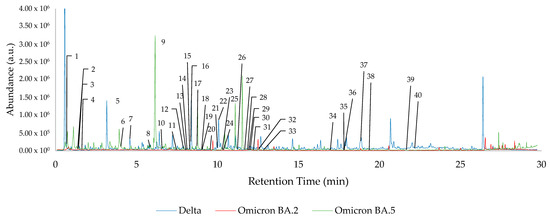
Figure 1.
Overlayed chromatograms of one patient per SARS-CoV-2 variant dataset, with all 40 features of interest highlighted.
2.8. Statistical Analysis
Models predicting the corresponding variant of the hand odor samples collected from COVID-19-positive patients were created using sparse partial least squares discriminant analysis (sPLS-DA). Three models were developed using sPLS-DA as the supervised machine learning algorithm, and each model was created using the log-10 transformed TIC peak areas of the 40 tracked features of interest. sPLS-DA modeling was performed using the “mixOmics” packages in R [19]. In all cases, the sPLS-DA models were informed by an equal number of samples for each variant. The lesser of the two compared variant sample sets was used as the defined class size, and the larger group was randomly sampled to provide an equal number of samples from each class. The models were cross-validated using a 5-fold cross-validation, which was repeated 200 times. This resulted in a random division of samples into 80% training set and 20% test set. The results of each of the 200 repetitions were used to calculate the mean accuracy of the model with a 95.0% confidence interval.
3. Results
Using sPLS-DA modeling, the sample subsets for the suspected Delta, Omicron BA.2, and Omicron BA.5 variants were compared to each other. As seen in Figure 1, the chromatograms corresponding to the patient hand odor from the different variant datasets are visually dissimilar; however, further in-depth data mining and statistical analysis was required for clear discrimination of the variants. Pre-processed, log-10 transformed TIC peak areas of the 40 features of interest were used to develop the following sPLS-DA models. Each model displays the class groupings of two COVID-19 variants (Delta, Omicron BA.2, or Omicron BA.5).
Figure 2 shows the sPLS-DA modeling of n = 20 positive Delta samples and n = 13 positive Omicron BA.2 samples. The ellipses between the two variant classes demonstrate some overlap in the 95.0% confidence interval ranges. This observed overlap is reflected in the performance of the cross-validated model, which correctly predicted the variant class with 71.2% (±1.0) accuracy. Within this model, an increased success rate was observed for correctly identifying Omicron BA.2 samples (79.6% ± 0.7) compared to the accuracy rate for Delta samples (62.8% ± 1.6).
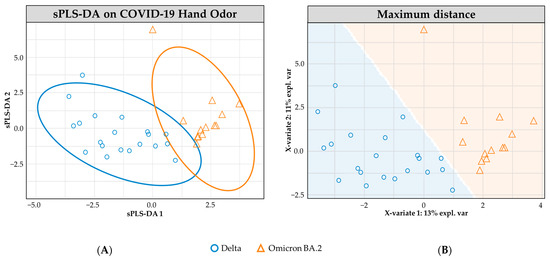
Figure 2.
sPLS-DA model for COVID-19 hand odor samples: (+) Delta vs. (+) Omicron BA.2. (A) Individual sample plot; ellipses = 95.0% CI. (B) Individual sample plot, background delineates class assignment.
Figure 3 demonstrates the sPLS-DA modeling of the positive COVID-19 Omicron BA.2 (n = 13) and Omicron BA.5 (n = 23) hand odor samples. The plotted model demonstrates little overlap between the two subclasses, using 95.0% confidence interval ellipses. The cross-validated model performed with an overall 78.5% (±0.8) accuracy rate. The model correctly predicted the class of Omicron BA.2 samples in 70.9% (±1.0) of occurrences and in 86.2% (±1.2) for the Omicron BA.5 samples.
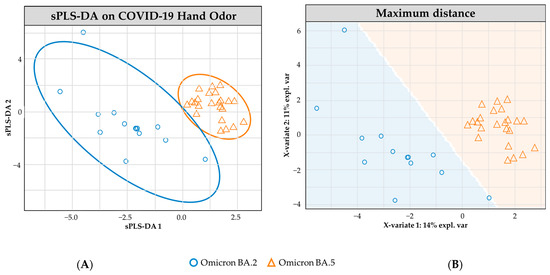
Figure 3.
sPLS-DA model for COVID-19 hand odor samples: (+) Omicron BA.2 vs. (+) Omicron BA.5. (A) Individual sample plot; ellipses = 95.0% CI. (B) Individual sample plot, background delineates class assignment.
Figure 4 shows the sPLS-DA modeling of n = 20 Delta- and n = 23 Omicron BA.5-positive COVID-19 hand odor samples. The model shows no overlap in the 95.0% confidence interval ellipses. The cross-validated model reflects a high performance in determining the variant class, with an overall accuracy rate of 95.2% (±0.4). The Delta-positive samples were correctly identified in 93.0% (±0.4) of cases, and the Omicron BA.5 samples were correctly identified in 97.4% (±0.5) of occurrences.
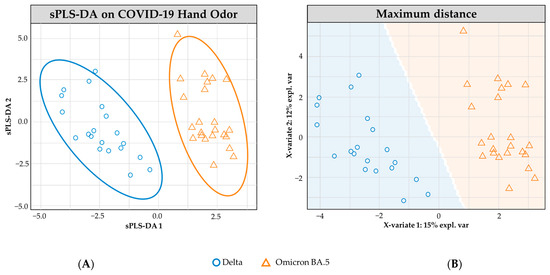
Figure 4.
sPLS-DA model for COVID-19 hand odor samples: (+) Delta vs. (+) Omicron BA.5. (A) Individual sample plot; ellipses= 95.0% CI. (B) Individual sample plot, background delineates class assignment.
4. Discussion
Human hand odor samples were collected from 56 SARS-CoV-2-positive patients in the PPMC Emergency Department in Philadelphia, PA. The SARS-CoV-2 variant in highest recorded abundance at the time of the sample’s collection was noted and attributed to that sample. The assignment of the SARS-CoV-2 variant was conducted using the CDC’s published reporting of SARS-CoV-2 presence within “Region 3” of the USA, which included Pennsylvania [1]. The samples were heated in sealed vials and sampled from the headspace using DVB/CAR/PDMS solid phase microextraction (SPME) fibers. The SPME fibers allowed for the capture and transfer of volatile organic compounds from the gaseous phase of the hand odor sample to the GC–MS. All 56 samples were analyzed in this manner, and the resulting GC–MS data files were processed in order to retrieve the chromatographic and mass spectral data relating to the 40 key features of interest.
The collected samples were used to develop models demonstrating the separability of the SARS-CoV-2 variants using the 40 key features of interest that can initially indicate whether a hand odor sample is collected from a SARS-CoV-2-positive or SARS-CoV-2-negative individual [16]. The collected samples were modeled to reveal the separability of the individual SARS-CoV-2 variants from one another as follows: (1) Delta vs. Omicron BA.2, (2) Omicron BA.2 vs. Omicron BA.5, and (3) Delta vs. Omicron BA.5. The models were constructed using the log-10 transformed TIC peak areas of the noted 40 features of interest, and the underlying theory of each model was the use of sparse partial least squares discriminant analysis (sPLS-DA). The 95.0% confidence interval ellipses that were formed by each sPLS-DA model demonstrate varying degrees of overlap, with Figure 2 (Delta vs. Omicron BA.2) reflecting the most overlap of the three models and Figure 4 (Delta vs. Omicron BA.5) demonstrating complete separability of the 95.0% confidence interval ellipses. The visual comparison of the three models is mirrored in their performance, with the greatest overall accuracy rate belonging to the Delta vs. Omicron BA.5 model (Figure 4); which demonstrated zero visual overlapping (the performance rate for this model indicated a 95.2% (±0.4) accuracy rate). The model with the second greatest overall accuracy rate was the Omicron BA.2 vs. Omicron BA.5 model (Figure 3), which demonstrated a 78.5% (±0.8) accuracy rate. Lastly, the Delta vs. Omicron BA.2 model (Figure 2) demonstrated a 71.2% (±1.0) accuracy rate. The performance rates of each model are broken down further in Table 2 to reflect the variant-specific accuracy rates.

Table 2.
sPLS-DA Model Performance for Variant Differentiation.
The accuracy rates achieved by these models indicate that the volatile organic compound expression exhibited by the Delta variant of the SARS-CoV-2-positive samples is more dissimilar from that of the Omicron BA.5 variant than the expression achieved by the Omicron BA.2 variant. Additionally, the moderate separability that still exists between the Delta and Omicron BA.2 variants indicates that the measurable VOC expressions that are associated with these disease variants are distinguishable from one another. There is an indication that each investigated SARS-CoV-2 variant causes a different detectable expression of disease state, which is critically important to acknowledge within the field of VOC-based diagnostic research.
Other researchers within the VOC-based diagnostics field are likewise investigating the use of human odor expression as a means of determining disease states. The presented findings strongly suggest that, as the SARS-CoV-2 virus changes and adapts, the odor expression that is induced also changes. This work demonstrates the need for continual training and tuning of odor-based diagnostic approaches. These approaches include (but are not limited to) the use of instrumental analysis, e-noses, and detection canines. The separability of variants also calls into question the viability of the techniques that were developed and trained solely on variants that are in low abundance in the population (Delta or earlier), as these variants induce notably different odor expressions in humans than the more recent Omicron variants. Such techniques that have not been reevaluated for their efficacy in correctly identifying currently prevailing SARS-CoV-2 variants stand to be assessed.
This work is bound by the limits of the existing gold-standard for determining SARS-CoV-2 status. There is no 100% accurate test for determining if an individual truly does have SARS-CoV-2. This study used PCR testing as its reference to determine the “ground truth;” however, there still exists the possibility that SARS-CoV-2-negative individuals were included in our sample set population. Additionally, this work was influenced by the limitations imposed by the make-up of the demographics of the patient population. The donor sample sets for the Delta, Omicron BA.2, and Omicron BA.5 variants did not equally reflect the donor characteristics of the study population as a whole (in terms of race, sex at birth, or age of donor (Table A1)). While these factors may contribute to increasing or decreasing the overlap between the odor expressions presented by each variant group, the features of interest that were monitored in this work were initially identified using a larger, more diverse sample population, with the aim of distinguishing SARS-CoV-2-positive and SARS-CoV-2-negative sample expressions [16]. Monitoring the same features of interest that elicit the separation of COVID-19-positive and COVID-19-negative samples is believed to have minimized the separability of the classes due to race, age, or sex factors, as these traits were considered during the initial feature selection procedure. These 40 features were initially chosen due to their ability to minimize within-class COVID-19-positive sample odor expression variability, taking inter-donor variation into account. Lastly, a general limitation of most pilot studies is the sample set itself. The researchers have formed these opinions based on the analysis of a sample set of n = 56 individual hand odor samples. Due to the nature of SARS-CoV-2 infections, the rates of positivity in Philadelphia, PA, and the willingness of patients to seek care and to participate in our study, a limited quantity of samples were collected for use in this work. Additionally, disparities between the variant sample set sizes were unavoidable, as the most prevalent variant in the area shifted multiple times throughout the sample collection period.
The studied variants were seen to display notably different odor expressions from one another, indicating that the evolution of the SARS-CoV-2 virus induces a change in one’s metabolic expression. This finding is critical to note within the field as it highlights the sensitive nature of this technique to detect not just disease state, but also the variant of the virus that is causing the disease. This raises the need for COVID-19 VOC-based detection methodologies to be informed by the currently prevalent SARS-CoV-2 variants. The highly distinguishable nature of the Delta vs. Omicron BA.5 variants suggests that a methodology built around the expression of the Delta variant may not recognize an Omicron BA.5-afflicted patient variant as being positive with SARS-CoV-2.
The researchers believe that the findings of this study can, and should, be expanded upon within the community to include VOC-based monitoring of collected and tested samples for the possible emergence of new variants. Such monitoring could be conducted through the use of a class inclusion/exclusion model, this type of approach could alert the researcher or practitioner to an out-of-class sample that is highly similar to the positive sample set. This expansion of VOC-based diagnostics could be combined with secondary rapid-antigen and/or PCR testing in order to provide further information about the positivity status, while the VOC analysis may be able to flag a potential new variant prior to the costly confirmatory analysis of genomic sequencing. The adoption of continuous monitoring of samples tested for SARS-CoV-2 for their degree of similarity may lead to an earlier recognition of emergent virus variants, as well as indication of the need for further training and tuning of models to reduce ambiguity about sample status (positive or negative). Overall, this study looked to demonstrate how the different metabolic expressions vary between distinct SARS-CoV-2 variants through a non-invasive analysis; it has highlighted how this may adversely affect current diagnostic techniques; and, finally, it has proposed new avenues to compensate for this limitation in the current methodologies.
5. Conclusions
Sparse partial least squares discriminant analysis was used to model the separability of the odor expression associated with the following three SARS-CoV-2 variants of interest: Delta, Omicron BA.2, and Omicron BA.5. This work revealed that VOCs from Delta and Omicron BA.5 samples were highly separable. The least separable variant combination was Delta vs. Omicron BA.2, and this model had moderate success in correctly predicting the variant class.
This work has demonstrated the separability of SARS-CoV-2 variants based upon the detectable odor expression exhibited by COVID-19-positive patients. This work stresses the importance of the continual training and evolution of VOC-based COVID-19 detection methodologies, as the expression of older variants is highly distinguishable from that of newer emerging variants. The ease of distinguishing these variants draws into question whether a model that is solely informed by older variants would correctly recognize emerging variants as being COVID-19-positive. It is vital to acknowledge this finding within the field, as it will guide further progress in developing non-invasive odor expression-based detection modalities, including the expansion upon analytical instrumentation-based approaches. This work has built upon previous findings, indicating the ability to determine the COVID-19 status of a patient using hand odor to state that the same sample can be used to predict the variant with high degrees of accuracy.
Author Contributions
Conceptualization, H.K.H., R.P. and K.G.F.; methodology, J.C.-C. and V.A.G.; software, V.A.G.; validation, J.C.-C. and V.A.G.; formal analysis, V.A.G. and J.C.-C.; data curation, V.A.G. and J.C.-C.; investigation, J.C.-C., V.A.G. and A.R.T.; resources, L.F., B.S.A. and K.G.F.; writing—original draft preparation, V.A.G. and J.C.-C.; writing—review and editing, V.A.G. and J.C.-C.; visualization, V.A.G. and J.C.-C.; supervision, H.K.H.; project administration, K.G.F. and A.T.C.J.; funding acquisition, K.G.F., R.P. and A.T.C.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the NIH National Center for Advancing Translational Sciences 1-U18-Tr-003775-01.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the University of Pennsylvania (IRB#843534; approved on 06/26/2020).
Data Availability Statement
The dataset associated with this study has been submitted to the National Institute of Health’s Scientific Data Sharing platform. Access to the dataset can be requested through https://sharing.nih.gov/. (accessed on 16 April 2023).
Acknowledgments
The researchers would like to thank the study participants for donating their time and hand odor samples. We would also like to thank the team at PARC-PPMC for their efforts on the groundwork in collecting and transferring the hand odor samples for instrumental analysis. The writing and preparation of this work was in part performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Demographic Breakdown of Subjects.
Table A1.
Demographic Breakdown of Subjects.
| Delta | Omicron BA.2 | Omicron BA.5 | ||
|---|---|---|---|---|
| Race | Black | 18 | 4 | 17 |
| White | - | 8 | 6 | |
| Hispanic/Latinx | 1 | 1 | - | |
| Unknown | 1 | - | - | |
| Sex at Birth | Female | 13 | 10 | 13 |
| Male | 7 | 3 | 10 | |
| Age | 18–24 | - | 5 | 1 |
| 25–30 | 3 | 1 | 4 | |
| 31–45 | 8 | 2 | 4 | |
| 46–60 | 4 | 3 | 6 | |
| 60+ | 5 | 2 | 8 |
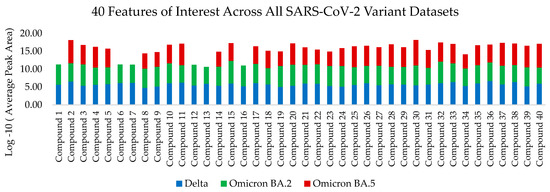
Figure A1.
Graphic representation of log-10 transformed average peak areas of all 40 features of interest for the Delta, Omicron BA.2 and Omicron BA.5 datasets.
References
- Centers for Disease Control and Prevention, CDC. COVID Data Tracker: Variant Proportions. Available online: https://covid.cdc.gov/covid-data-tracker/#variant-proportions (accessed on 12 August 2022).
- Fernandes, Q.; Inchakalody, V.P.; Merhi, M.; Mestiri, S.; Taib, N.; Moustafa Abo El-Ella, D.; Bedhiafi, T.; Raza, A.; Al-Zaidan, L.; Mohsen, M.O.; et al. Emerging COVID-19 Variants and Their Impact on SARS-CoV-2 Diagnosis, Therapeutics and Vaccines. Ann. Med. 2022, 54, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html#concern (accessed on 8 December 2022).
- Ninomiya, M.; Ueno, Y.; Funayama, R.; Nagashima, T.; Nishida, Y.; Kondo, Y.; Inoue, J.; Kakazu, E.; Kimura, O.; Nakayama, K.; et al. Use of Illumina Deep Sequencing Technology To Differentiate Hepatitis C Virus Variants. J. Clin. Microbiol. 2012, 50, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Hokynar, K.; Norja, P.; Laitinen, H.; Palomäki, P.; Garbarg-Chenon, A.; Ranki, A.; Hedman, K.; Söderlund-Venermo, M. Detection and Differentiation of Human Parvovirus Variants by Commercial Quantitative Real-Time PCR Tests. J. Clin. Microbiol. 2004, 42, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.M.; Jackwood, M.W.; Gelb, J. Differentiation of Infectious Bronchitis Virus Serotypes Using Polymerase Chain Reaction and Restriction Fragment Length Polymorphism Analysis. Avian Dis. 1993, 37, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Vo, V.; Tillett, R.L.; Papp, K.; Shen, S.; Gu, R.; Gorzalski, A.; Siao, D.; Markland, R.; Chang, C.L.; Baker, H.; et al. Use of Wastewater Surveillance for Early Detection of Alpha and Epsilon SARS-CoV-2 Variants of Concern and Estimation of Overall COVID-19 Infection Burden. Sci. Total Environ. 2022, 835, 155410. [Google Scholar] [CrossRef] [PubMed]
- Erster, O.; Mendelson, E.; Levy, V.; Kabat, A.; Mannasse, B.; Asraf, H.; Azar, R.; Ali, Y.; Shirazi, R.; Bucris, E.; et al. Rapid and High-Throughput Reverse Transcriptase Quantitative PCR (RT-QPCR) Assay for Identification and Differentiation between SARS-CoV-2 Variants B.1.1.7 and B.1.351. Microbiol. Spectr. 2021, 9, e00506-21. [Google Scholar] [CrossRef] [PubMed]
- Roche. Roche Launches COVID-19 Test That Detects and Differentiates the Emerging Variant of Interest BA.2.75. Available online: https://www.prnewswire.com/news-releases/roche-launches-covid-19-test-that-detects-and-differentiates-the-emerging-variant-of-interest-ba2-75--301609342.html (accessed on 2 September 2022).
- Mendel, J.; Frank, K.; Edlin, L.; Hall, K.; Webb, D.; Mills, J.; Holness, H.K.; Furton, K.G.; Mills, D.E. Preliminary Accuracy of COVID-19 Odor Detection by Canines and HS-SPME-GC-MS Using Exhaled Breath Samples. Forensic Sci. Int. 2021, 3, 100155. [Google Scholar] [CrossRef] [PubMed]
- Ruszkiewicz, D.M.; Sanders, D.; O’Brien, R.; Hempel, F.; Reed, M.J.; Riepe, A.C.; Bailie, K.; Brodrick, E.; Darnley, K.; Ellerkmann, R.; et al. Diagnosis of COVID-19 by Analysis of Breath with Gas Chromatography-Ion Mobility Spectrometry—A Feasibility Study. EClinicalMedicine 2020, 29–30, 100609. [Google Scholar] [CrossRef] [PubMed]
- Gokool, V.A.; Crespo-Cajigas, J.; Mallikarjun, A.; Collins, A.; Kane, S.A.; Plymouth, V.; Nguyen, E.; Abella, B.S.; Holness, H.K.; Furton, K.G.; et al. The Use of Biological Sensors and Instrumental Analysis to Discriminate COVID-19 Odor Signatures. Biosensors 2022, 12, 1003. [Google Scholar] [CrossRef] [PubMed]
- Curran, A.M.; Rabin, S.I.; Furton, K.G. Analysis of the Uniqueness and Persistence of Human Scent. Forensic Sci. Commun. 2005, 7, 1–12. [Google Scholar]
- Shirasu, M.; Touhara, K. The Scent of Disease: Volatile Organic Compounds of the Human Body Related to Disease and Disorder. J. Biochem. 2011, 150, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Woollam, M.; Angarita-Rivera, P.; Siegel, A.P.; Kalra, V.; Kapoor, R.; Agarwal, M. Exhaled VOCs Can Discriminate Subjects with COVID-19 from Healthy Controls. J. Breath Res. 2022, 16, 036002. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Cajigas, J.; Gokool, V.A.; Ramírez Torres, A.; Forsythe, L.; Abella, B.S.; Holness, H.K.; Johnson, A.T.C.; Postrel, R.; Furton, K.G. Investigating the Use of SARS-CoV-2 (COVID-19) Odor Expression as a Non-Invasive Diagnostic Tool—Pilot Study. Diagnostics 2023, 13, 707. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. cobas SARS-CoV-2—Instructions For Use. Available online: https://www.fda.gov/media/136049/download (accessed on 14 January 2023).
- Food and Drug Administration. BINAXNOW COVID-19 AG CARD (PN 195-000)—INSTRUCTIONS FOR USE. Available online: https://www.fda.gov/media/141570/download (accessed on 14 January 2023).
- Le Cao, K.-A.; Rohart, F.; Gonzalez, I.; Dejean, S.; Gautier, B.; Bartolo, F.; Monget, P.; Coquery, J.; Yao, F.; Liquet, B. mixOmics: Omics Data Integration Project. R Package Version 6.1.1. Available online: https://CRAN.R-project.org/package=mixOmics (accessed on 11 August 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).