Understanding Macrophage Complexity in Metabolic Dysfunction-Associated Steatotic Liver Disease: Transitioning from the M1/M2 Paradigm to Spatial Dynamics
Abstract
1. Introduction
2. General Overview of Macrophages
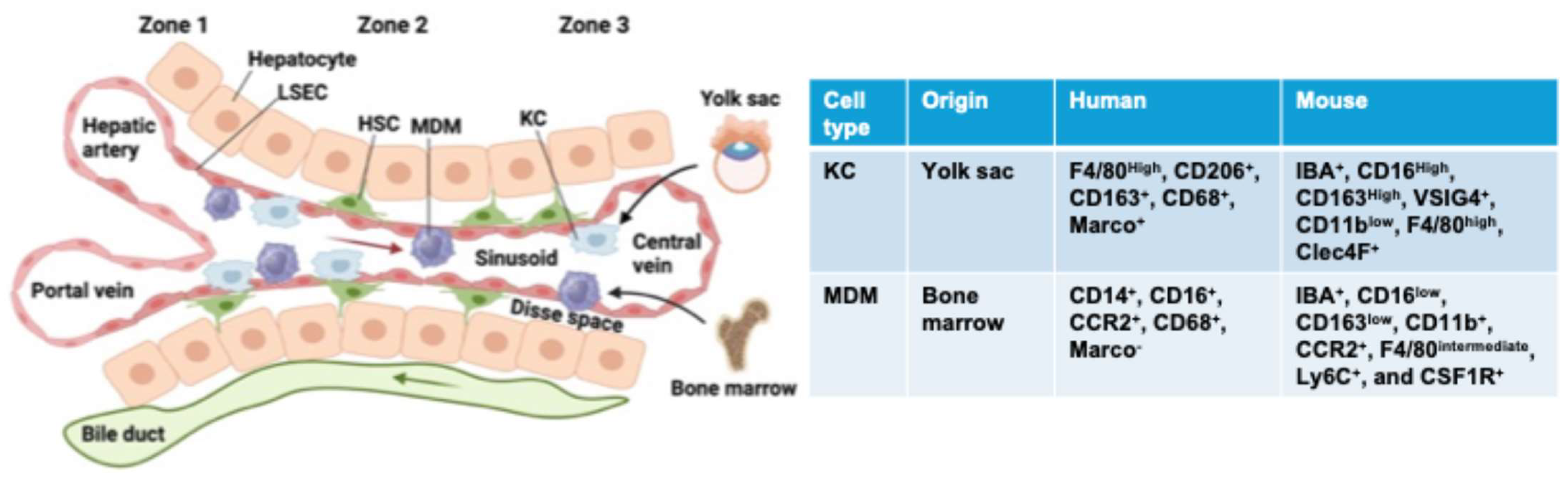
3. Hepatic Macrophages: Type, Origin, and Function
4. Macrophage Accumulation in MASLD: Insights from Animal Models and Human Studies
5. Stimuli Trigger Hepatic Macrophage Activation during MASLD Development
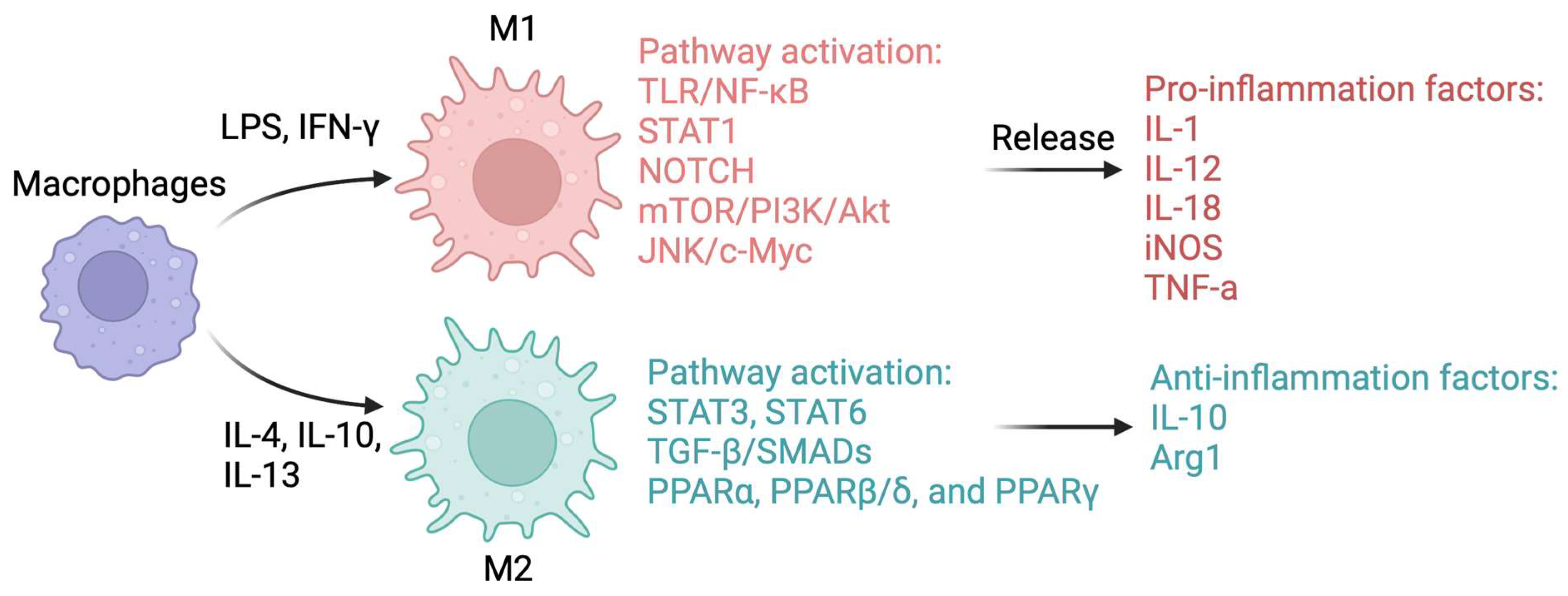
6. Classical M1/M2 Macrophage Paradigm in MASLD Development
6.1. Mechanisms Control Macrophage Polarization
6.1.1. TLR and NF-κB
6.1.2. Signal Transducer and Activator of Transcription (STAT)
6.1.3. Transforming Growth Factor Beta (TGF-β) and Suppressor of Mothers against Decapentaplegic (SMAD)
6.1.4. Peroxisome Proliferator-Activated Receptors (PPARs)
6.1.5. MicroRNAs (miRNAs) and Other Mechanisms
6.2. Macrophage Polarization in Early Stage of MASLD
6.3. Macrophage Polarization in Advanced Stage of MASLD
7. Revealing the Dynamic Landscape of Hepatic Macrophages in MASLD: Heterogeneity and Plasticity
8. Unraveling the Complexity of Hepatic Macrophages in MASLD: Insights into Spatial Dynamics
9. Targeting Macrophages for the Treatment of MASLD
10. Conclusions and Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Rinella, M.E.; Sookoian, S. From NAFLD to MASLD: Updated naming and diagnosis criteria for fatty liver disease. J. Lipid Res. 2024, 65, 100485. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Estes, C.; Anstee, Q.M.; Arias-Loste, M.T.; Bantel, H.; Bellentani, S.; Caballeria, J.; Colombo, M.; Craxi, A.; Crespo, J.; Day, C.P.; et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016–2030. J. Hepatol. 2018, 69, 896–904. [Google Scholar] [CrossRef]
- Younossi, Z.; Tacke, F.; Arrese, M.; Chander Sharma, B.; Mostafa, I.; Bugianesi, E.; Wai-Sun Wong, V.; Yilmaz, Y.; George, J.; Fan, J.; et al. Global Perspectives on Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Hepatology 2019, 69, 2672–2682. [Google Scholar] [CrossRef]
- Schuppan, D.; Surabattula, R.; Wang, X.Y. Determinants of fibrosis progression and regression in NASH. J. Hepatol. 2018, 68, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.D.; Stengel, J.; Asike, M.I.; Torres, D.M.; Shaw, J.; Contreras, M.; Landt, C.L.; Harrison, S.A. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: A prospective study. Gastroenterology 2011, 140, 124–131. [Google Scholar] [CrossRef]
- Sayiner, M.; Koenig, A.; Henry, L.; Younossi, Z.M. Epidemiology of Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis in the United States and the Rest of the World. Clin. Liver Dis. 2016, 20, 205–214. [Google Scholar] [CrossRef]
- Walker, R.W.; Le, K.A.; Davis, J.; Alderete, T.L.; Cherry, R.; Lebel, S.; Goran, M.I. High rates of fructose malabsorption are associated with reduced liver fat in obese African Americans. J. Am. Coll. Nutr. 2012, 31, 369–374. [Google Scholar] [CrossRef]
- Wong, R.J.; Aguilar, M.; Cheung, R.; Perumpail, R.B.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015, 148, 547–555. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Blissett, D.; Blissett, R.; Henry, L.; Stepanova, M.; Younossi, Y.; Racila, A.; Hunt, S.; Beckerman, R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 2016, 64, 1577–1586. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Tampi, R.; Priyadarshini, M.; Nader, F.; Younossi, I.M.; Racila, A. Burden of Illness and Economic Model for Patients with Nonalcoholic Steatohepatitis in the United States. Hepatology 2019, 69, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Magee, N.; Zou, A.; Zhang, Y. Pathogenesis of Nonalcoholic Steatohepatitis: Interactions between Liver Parenchymal and Nonparenchymal Cells. Biomed. Res. Int. 2016, 2016, 5170402. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P.; James, O.F. Steatohepatitis: A tale of two “hits”? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P. From fat to inflammation. Gastroenterology 2006, 130, 207–210. [Google Scholar] [CrossRef]
- Jou, J.; Choi, S.S.; Diehl, A.M. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin. Liver Dis. 2008, 28, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Dowman, J.K.; Tomlinson, J.W.; Newsome, P.N. Pathogenesis of non-alcoholic fatty liver disease. QJM 2010, 103, 71–83. [Google Scholar] [CrossRef]
- Roskams, T.; Yang, S.Q.; Koteish, A.; Durnez, A.; DeVos, R.; Huang, X.; Achten, R.; Verslype, C.; Diehl, A.M. Oxidative stress and oval cell accumulation in mice and humans with alcoholic and nonalcoholic fatty liver disease. Am. J. Pathol. 2003, 163, 1301–1311. [Google Scholar] [CrossRef]
- Lopez, B.G.; Tsai, M.S.; Baratta, J.L.; Longmuir, K.J.; Robertson, R.T. Characterization of Kupffer cells in livers of developing mice. Comp. Hepatol. 2011, 10, 2. [Google Scholar] [CrossRef]
- Tacke, F.; Zimmermann, H.W. Macrophage heterogeneity in liver injury and fibrosis. J. Hepatol. 2014, 60, 1090–1096. [Google Scholar] [CrossRef]
- Arrese, M.; Cabrera, D.; Kalergis, A.M.; Feldstein, A.E. Innate Immunity and Inflammation in NAFLD/NASH. Dig. Dis. Sci. 2016, 61, 1294–1303. [Google Scholar] [CrossRef]
- Sutti, S.; Bruzzi, S.; Albano, E. The role of immune mechanisms in alcoholic and nonalcoholic steatohepatitis: A 2015 update. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Elie Metchnikoff: Father of natural immunity. Eur. J. Immunol. 2008, 38, 3257–3264 PMID: 19039772. [Google Scholar] [CrossRef] [PubMed]
- van Furth, R.; Cohn, Z.A.; Hirsch, J.G.; Humphrey, J.H.; Spector, W.G.; Langevoort, H.L. The mononuclear phagocyte system: A new classification of macrophages, monocytes, and their precursor cells. Bull. World Health Organ. 1972, 46, 845–852. [Google Scholar] [PubMed]
- Hume, D.A. The mononuclear phagocyte system. Curr. Opin. Immunol. 2006, 18, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Gautier, E.L.; Shay, T.; Miller, J.; Greter, M.; Jakubzick, C.; Ivanov, S.; Helft, J.; Chow, A.; Elpek, K.G.; Gordonov, S.; et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012, 13, 1118–1128. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- Schulz, C.; Gomez Perdiguero, E.; Chorro, L.; Szabo-Rogers, H.; Cagnard, N.; Kierdorf, K.; Prinz, M.; Wu, B.; Jacobsen, S.E.; Pollard, J.W.; et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012, 336, 86–90. [Google Scholar] [CrossRef]
- Yona, S.; Kim, K.W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013, 38, 79–91. [Google Scholar] [CrossRef]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef]
- Ley, K.; Pramod, A.B.; Croft, M.; Ravichandran, K.S.; Ting, J.P. How Mouse Macrophages Sense What Is Going On. Front. Immunol. 2016, 7, 204. [Google Scholar] [CrossRef]
- Gordon, S. Phagocytosis: The Legacy of Metchnikoff. Cell 2016, 166, 1065–1068. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Park, M.D.; Silvin, A.; Ginhoux, F.; Merad, M. Macrophages in health and disease. Cell 2022, 185, 4259–4279. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Giladi, A.; Gorki, A.D.; Solodkin, D.G.; Zada, M.; Hladik, A.; Miklosi, A.; Salame, T.M.; Halpern, K.B.; David, E.; et al. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell 2018, 175, 1031–1044.e18. [Google Scholar] [CrossRef]
- Amit, I.; Winter, D.R.; Jung, S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat. Immunol. 2016, 17, 18–25. [Google Scholar] [CrossRef]
- Gosselin, D.; Link, V.M.; Romanoski, C.E.; Fonseca, G.J.; Eichenfield, D.Z.; Spann, N.J.; Stender, J.D.; Chun, H.B.; Garner, H.; Geissmann, F.; et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014, 159, 1327–1340. [Google Scholar] [CrossRef]
- Lavin, Y.; Merad, M. Macrophages: Gatekeepers of tissue integrity. Cancer Immunol. Res. 2013, 1, 201–209. [Google Scholar] [CrossRef]
- Pollard, J.W. Trophic macrophages in development and disease. Nat. Rev. Immunol. 2009, 9, 259–270. [Google Scholar] [CrossRef]
- Van Nguyen, A.; Pollard, J.W. Colony stimulating factor-1 is required to recruit macrophages into the mammary gland to facilitate mammary ductal outgrowth. Dev. Biol. 2002, 247, 11–25. [Google Scholar] [CrossRef]
- Banaei-Bouchareb, L.; Gouon-Evans, V.; Samara-Boustani, D.; Castellotti, M.C.; Czernichow, P.; Pollard, J.W.; Polak, M. Insulin cell mass is altered in Csf1op/Csf1op macrophage-deficient mice. J. Leukoc. Biol. 2004, 76, 359–367. [Google Scholar] [CrossRef]
- Wiktor-Jedrzejczak, W.; Bartocci, A.; Ferrante, A.W., Jr.; Ahmed-Ansari, A.; Sell, K.W.; Pollard, J.W.; Stanley, E.R. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc. Natl. Acad. Sci. USA 1990, 87, 4828–4832. [Google Scholar] [CrossRef] [PubMed]
- Chasis, J.A.; Mohandas, N. Erythroblastic islands: Niches for erythropoiesis. Blood 2008, 112, 470–478. [Google Scholar] [CrossRef]
- Sadahira, Y.; Yoshino, T.; Monobe, Y. Very late activation antigen 4-vascular cell adhesion molecule 1 interaction is involved in the formation of erythroblastic islands. J. Exp. Med. 1995, 181, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Bessis, M.C.; Breton-Gorius, J. Iron metabolism in the bone marrow as seen by electron microscopy: A critical review. Blood 1962, 19, 635–663. [Google Scholar] [CrossRef] [PubMed]
- Skutelsky, E.; Danon, D. On the expulsion of the erythroid nucleus and its phagocytosis. Anat. Rec. 1972, 173, 123–126. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System during Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef]
- Erblich, B.; Zhu, L.; Etgen, A.M.; Dobrenis, K.; Pollard, J.W. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS ONE 2011, 6, e26317. [Google Scholar] [CrossRef]
- Whitsett, J.A.; Wert, S.E.; Weaver, T.E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 2010, 61, 105–119. [Google Scholar] [CrossRef]
- Wake, K. Karl Wilhelm Kupffer and His Contributions to Modern Hepatology. Comp. Hepatol. 2004, 3 (Suppl. S1), S2. [Google Scholar] [CrossRef][Green Version]
- Bilzer, M.; Roggel, F.; Gerbes, A.L. Role of Kupffer cells in host defense and liver disease. Liver Int. 2006, 26, 1175–1186. [Google Scholar] [CrossRef]
- Bouwens, L.; Baekeland, M.; De Zanger, R.; Wisse, E. Quantitation, tissue distribution and proliferation kinetics of Kupffer cells in normal rat liver. Hepatology 1986, 6, 718–722. [Google Scholar] [CrossRef] [PubMed]
- MacPhee, P.J.; Schmidt, E.E.; Groom, A.C. Evidence for Kupffer cell migration along liver sinusoids, from high-resolution in vivo microscopy. Am. J. Physiol. 1992, 263, G17–G23. [Google Scholar] [CrossRef]
- Wen, Y.; Lambrecht, J.; Ju, C.; Tacke, F. Hepatic macrophages in liver homeostasis and diseases-diversity, plasticity and therapeutic opportunities. Cell. Mol. Immunol. 2021, 18, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Krenkel, O.; Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 2017, 17, 306–321. [Google Scholar] [CrossRef]
- Zigmond, E.; Samia-Grinberg, S.; Pasmanik-Chor, M.; Brazowski, E.; Shibolet, O.; Halpern, Z.; Varol, C. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J. Immunol. 2014, 193, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Scott, C.L.; Zheng, F.; De Baetselier, P.; Martens, L.; Saeys, Y.; De Prijck, S.; Lippens, S.; Abels, C.; Schoonooghe, S.; Raes, G.; et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 2016, 7, 10321. [Google Scholar] [CrossRef]
- Kim, K.W.; Zhang, N.; Choi, K.; Randolph, G.J. Homegrown Macrophages. Immunity 2016, 45, 468–470. [Google Scholar] [CrossRef][Green Version]
- Hoeffel, G.; Chen, J.; Lavin, Y.; Low, D.; Almeida, F.F.; See, P.; Beaudin, A.E.; Lum, J.; Low, I.; Forsberg, E.C.; et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 2015, 42, 665–678. [Google Scholar] [CrossRef]
- Gomez Perdiguero, E.; Klapproth, K.; Schulz, C.; Busch, K.; Azzoni, E.; Crozet, L.; Garner, H.; Trouillet, C.; de Bruijn, M.F.; Geissmann, F.; et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518, 547–551. [Google Scholar] [CrossRef]
- Bouwens, L.; Knook, D.L.; Wisse, E. Local proliferation and extrahepatic recruitment of liver macrophages (Kupffer cells) in partial-body irradiated rats. J. Leukoc. Biol. 1986, 39, 687–697. [Google Scholar] [CrossRef]
- Wacker, H.H.; Radzun, H.J.; Parwaresch, M.R. Kinetics of Kupffer cells as shown by parabiosis and combined autoradiographic/immunohistochemical analysis. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1986, 51, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Naito, M.; Moriyama, H.; Umezu, H.; Matsuo, H.; Kiwada, H.; Arakawa, M. Repopulation of murine Kupffer cells after intravenous administration of liposome-encapsulated dichloromethylene diphosphonate. Am. J. Pathol. 1996, 149, 1271–1286. [Google Scholar]
- Fogg, D.K.; Sibon, C.; Miled, C.; Jung, S.; Aucouturier, P.; Littman, D.R.; Cumano, A.; Geissmann, F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 2006, 311, 83–87. [Google Scholar] [CrossRef] [PubMed]
- MacParland, S.A.; Liu, J.C.; Ma, X.Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, S.; Liu, Y.; He, X.; Qu, M.; Xu, G.; Wang, H.; Huang, M.; Pan, J.; Liu, Z.; et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020, 6, 22. [Google Scholar] [CrossRef]
- Aizarani, N.; Saviano, A.; Sagar; Mailly, L.; Durand, S.; Herman, J.S.; Pessaux, P.; Baumert, T.F.; Grun, D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019, 572, 199–204. [Google Scholar] [CrossRef]
- Matchett, K.P.; Wilson-Kanamori, J.R.; Portman, J.R.; Kapourani, C.A.; Fercoq, F.; May, S.; Zajdel, E.; Beltran, M.; Sutherland, E.F.; Mackey, J.B.G.; et al. Multimodal decoding of human liver regeneration. Nature 2024, 630, 158–165. [Google Scholar] [CrossRef]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Popescu, D.M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nature 2019, 574, 365–371. [Google Scholar] [CrossRef]
- Pallett, L.J.; Burton, A.R.; Amin, O.E.; Rodriguez-Tajes, S.; Patel, A.A.; Zakeri, N.; Jeffery-Smith, A.; Swadling, L.; Schmidt, N.M.; Baiges, A.; et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J. Exp. Med. 2020, 217, e20200050. [Google Scholar] [CrossRef]
- Heymann, F.; Peusquens, J.; Ludwig-Portugall, I.; Kohlhepp, M.; Ergen, C.; Niemietz, P.; Martin, C.; van Rooijen, N.; Ochando, J.C.; Randolph, G.J.; et al. Liver inflammation abrogates immunological tolerance induced by Kupffer cells. Hepatology 2015, 62, 279–291. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Cheng, L.; Kedl, R.M.; Ju, C. Mechanism of T cell tolerance induction by murine hepatic Kupffer cells. Hepatology 2008, 48, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Magee, N.; Ahamed, F.; Eppler, N.; Jones, E.; Ghosh, P.; He, L.; Zhang, Y. Hepatic transcriptome profiling reveals early signatures associated with disease transition from non-alcoholic steatosis to steatohepatitis. Liver Res. 2022, 6, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Obstfeld, A.E.; Sugaru, E.; Thearle, M.; Francisco, A.M.; Gayet, C.; Ginsberg, H.N.; Ables, E.V.; Ferrante, A.W., Jr. C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes 2010, 59, 916–925. [Google Scholar] [CrossRef]
- Ni, Y.; Nagashimada, M.; Zhuge, F.; Zhan, L.; Nagata, N.; Tsutsui, A.; Nakanuma, Y.; Kaneko, S.; Ota, T. Astaxanthin prevents and reverses diet-induced insulin resistance and steatohepatitis in mice: A comparison with vitamin E. Sci. Rep. 2015, 5, 17192. [Google Scholar] [CrossRef]
- Devisscher, L.; Scott, C.L.; Lefere, S.; Raevens, S.; Bogaerts, E.; Paridaens, A.; Verhelst, X.; Geerts, A.; Guilliams, M.; Van Vlierberghe, H. Non-alcoholic steatohepatitis induces transient changes within the liver macrophage pool. Cell. Immunol. 2017, 322, 74–83. [Google Scholar] [CrossRef]
- Lefere, S.; Degroote, H.; Van Vlierberghe, H.; Devisscher, L. Unveiling the depletion of Kupffer cells in experimental hepatocarcinogenesis through liver macrophage subtype-specific markers. J. Hepatol. 2019, 71, 631–633. [Google Scholar] [CrossRef]
- Tosello-Trampont, A.C.; Landes, S.G.; Nguyen, V.; Novobrantseva, T.I.; Hahn, Y.S. Kuppfer cells trigger nonalcoholic steatohepatitis development in diet-induced mouse model through tumor necrosis factor-alpha production. J. Biol. Chem. 2012, 287, 40161–40172. [Google Scholar] [CrossRef]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef]
- Singh, S.; Anshita, D.; Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol. 2021, 101, 107598. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, Y. CCL2-CCR2 signaling axis in obesity and metabolic diseases. J. Cell. Physiol. 2024, 239, e31192. [Google Scholar] [CrossRef] [PubMed]
- Morinaga, H.; Mayoral, R.; Heinrichsdorff, J.; Osborn, O.; Franck, N.; Hah, N.; Walenta, E.; Bandyopadhyay, G.; Pessentheiner, A.R.; Chi, T.J.; et al. Characterization of distinct subpopulations of hepatic macrophages in HFD/obese mice. Diabetes 2015, 64, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Krenkel, O.; Puengel, T.; Govaere, O.; Abdallah, A.T.; Mossanen, J.C.; Kohlhepp, M.; Liepelt, A.; Lefebvre, E.; Luedde, T.; Hellerbrand, C.; et al. Therapeutic inhibition of inflammatory monocyte recruitment reduces steatohepatitis and liver fibrosis. Hepatology 2018, 67, 1270–1283. [Google Scholar] [CrossRef]
- Park, J.W.; Jeong, G.; Kim, S.J.; Kim, M.K.; Park, S.M. Predictors reflecting the pathological severity of non-alcoholic fatty liver disease: Comprehensive study of clinical and immunohistochemical findings in younger Asian patients. J. Gastroenterol. Hepatol. 2007, 22, 491–497. [Google Scholar] [CrossRef]
- Lotowska, J.M.; Sobaniec-Lotowska, M.E.; Lebensztejn, D.M. The role of Kupffer cells in the morphogenesis of nonalcoholic steatohepatitis—Ultrastructural findings. The first report in pediatric patients. Scand. J. Gastroenterol. 2013, 48, 352–357. [Google Scholar] [CrossRef]
- Gadd, V.L.; Skoien, R.; Powell, E.E.; Fagan, K.J.; Winterford, C.; Horsfall, L.; Irvine, K.; Clouston, A.D. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology 2014, 59, 1393–1405. [Google Scholar] [CrossRef]
- Kazankov, K.; Jorgensen, S.M.D.; Thomsen, K.L.; Moller, H.J.; Vilstrup, H.; George, J.; Schuppan, D.; Gronbaek, H. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Kato, H.; Suganami, T.; Konuma, K.; Marumoto, Y.; Terai, S.; Sakugawa, H.; Kanai, S.; Hamaguchi, M.; Fukaishi, T.; et al. Hepatic crown-like structure: A unique histological feature in non-alcoholic steatohepatitis in mice and humans. PLoS ONE 2013, 8, e82163. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology 2010, 52, 774–788. [Google Scholar] [CrossRef]
- Pan, X.; Wang, P.; Luo, J.; Wang, Z.; Song, Y.; Ye, J.; Hou, X. Adipogenic changes of hepatocytes in a high-fat diet-induced fatty liver mice model and non-alcoholic fatty liver disease patients. Endocrine 2015, 48, 834–847. [Google Scholar] [CrossRef]
- Lee, J.Y.; Sohn, K.H.; Rhee, S.H.; Hwang, D. Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J. Biol. Chem. 2001, 276, 16683–16689. [Google Scholar] [CrossRef]
- Snodgrass, R.G.; Huang, S.; Choi, I.W.; Rutledge, J.C.; Hwang, D.H. Inflammasome-mediated secretion of IL-1beta in human monocytes through TLR2 activation; modulation by dietary fatty acids. J. Immunol. 2013, 191, 4337–4347. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Jeong, J.M.; Kim, S.J.; Seo, W.; Kim, M.H.; Choi, W.M.; Yoo, W.; Lee, J.H.; Shim, Y.R.; Yi, H.S.; et al. Pro-inflammatory hepatic macrophages generate ROS through NADPH oxidase 2 via endocytosis of monomeric TLR4-MD2 complex. Nat. Commun. 2017, 8, 2247. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, R.; Moreno-Fernandez, M.E.; Giles, D.A.; Cappelletti, M.; Stankiewicz, T.E.; Chan, C.C.; Divanovic, S. Nicotinamide adenine dinucleotide phosphate (reduced) oxidase 2 modulates inflammatory vigor during nonalcoholic fatty liver disease progression in mice. Hepatol. Commun. 2018, 2, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Del Ben, M.; Polimeni, L.; Carnevale, R.; Bartimoccia, S.; Nocella, C.; Baratta, F.; Loffredo, L.; Pignatelli, P.; Violi, F.; Angelico, F. NOX2-generated oxidative stress is associated with severity of ultrasound liver steatosis in patients with non-alcoholic fatty liver disease. BMC Gastroenterol. 2014, 14, 81. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog. Lipid Res. 2013, 52, 175–191. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Haigh, W.G.; Thorning, D.; Savard, C. Hepatic cholesterol crystals and crown-like structures distinguish NASH from simple steatosis. J. Lipid Res. 2013, 54, 1326–1334. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Subramanian, S.; Chait, A.; Haigh, W.G.; Yeh, M.M.; Farrell, G.C.; Lee, S.P.; Savard, C. Cholesterol crystallization within hepatocyte lipid droplets and its role in murine NASH. J. Lipid Res. 2017, 58, 1067–1079. [Google Scholar] [CrossRef]
- Duewell, P.; Kono, H.; Rayner, K.J.; Sirois, C.M.; Vladimer, G.; Bauernfeind, F.G.; Abela, G.S.; Franchi, L.; Nunez, G.; Schnurr, M.; et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 2010, 464, 1357–1361. [Google Scholar] [CrossRef]
- Rajamaki, K.; Lappalainen, J.; Oorni, K.; Valimaki, E.; Matikainen, S.; Kovanen, P.T.; Eklund, K.K. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: A novel link between cholesterol metabolism and inflammation. PLoS ONE 2010, 5, e11765. [Google Scholar] [CrossRef]
- Miller, Y.I.; Choi, S.H.; Wiesner, P.; Fang, L.; Harkewicz, R.; Hartvigsen, K.; Boullier, A.; Gonen, A.; Diehl, C.J.; Que, X.; et al. Oxidation-specific epitopes are danger-associated molecular patterns recognized by pattern recognition receptors of innate immunity. Circ. Res. 2011, 108, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Canbay, A.; Feldstein, A.E.; Higuchi, H.; Werneburg, N.; Grambihler, A.; Bronk, S.F.; Gores, G.J. Kupffer cell engulfment of apoptotic bodies stimulates death ligand and cytokine expression. Hepatology 2003, 38, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Hirsova, P.; Ibrahim, S.H.; Krishnan, A.; Verma, V.K.; Bronk, S.F.; Werneburg, N.W.; Charlton, M.R.; Shah, V.H.; Malhi, H.; Gores, G.J. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles from Hepatocytes. Gastroenterology 2016, 150, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, Z.; Xia, M.; Salas, S.S.; Ospina, J.A.; Buist-Homan, M.; Harmsen, M.C.; Moshage, H. Extracellular vesicles derived from liver sinusoidal endothelial cells inhibit the activation of hepatic stellate cells and Kupffer cells in vitro. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 167020. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed]
- Frasinariu, O.E.; Ceccarelli, S.; Alisi, A.; Moraru, E.; Nobili, V. Gut-liver axis and fibrosis in nonalcoholic fatty liver disease: An input for novel therapies. Dig. Liver Dis. 2013, 45, 543–551. [Google Scholar] [CrossRef]
- Budick-Harmelin, N.; Dudas, J.; Demuth, J.; Madar, Z.; Ramadori, G.; Tirosh, O. Triglycerides potentiate the inflammatory response in rat Kupffer cells. Antioxid. Redox Signal. 2008, 10, 2009–2022. [Google Scholar] [CrossRef]
- Leroux, A.; Ferrere, G.; Godie, V.; Cailleux, F.; Renoud, M.L.; Gaudin, F.; Naveau, S.; Prevot, S.; Makhzami, S.; Perlemuter, G.; et al. Toxic lipids stored by Kupffer cells correlates with their pro-inflammatory phenotype at an early stage of steatohepatitis. J. Hepatol. 2012, 57, 141–149. [Google Scholar] [CrossRef]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Coffman, R.L. TH1 and TH2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 1989, 7, 145–173. [Google Scholar] [CrossRef]
- Poltorak, A.; Smirnova, I.; He, X.; Liu, M.Y.; Van Huffel, C.; McNally, O.; Birdwell, D.; Alejos, E.; Silva, M.; Du, X.; et al. Genetic and physical mapping of the Lps locus: Identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol. Dis. 1998, 24, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Ju, Z.; Ragab, A.A.; Lundback, P.; Long, W.; Valdes-Ferrer, S.I.; He, M.; Pribis, J.P.; Li, J.; et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J. Exp. Med. 2015, 212, 5–14. [Google Scholar] [CrossRef]
- Jiang, D.; Liang, J.; Fan, J.; Yu, S.; Chen, S.; Luo, Y.; Prestwich, G.D.; Mascarenhas, M.M.; Garg, H.G.; Quinn, D.A.; et al. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat. Med. 2005, 11, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.X.; Tang, L.; Fu, Y.M.; Wang, Y.; Han, Z.H.; Meng, J.G. Paralemmin-3 contributes to lipopolysaccharide-induced inflammatory response and is involved in lipopolysaccharide-Toll-like receptor-4 signaling in alveolar macrophages. Int. J. Mol. Med. 2017, 40, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
- Suganami, T.; Mieda, T.; Itoh, M.; Shimoda, Y.; Kamei, Y.; Ogawa, Y. Attenuation of obesity-induced adipose tissue inflammation in C3H/HeJ mice carrying a Toll-like receptor 4 mutation. Biochem. Biophys. Res. Commun. 2007, 354, 45–49. [Google Scholar] [CrossRef]
- Csak, T.; Velayudham, A.; Hritz, I.; Petrasek, J.; Levin, I.; Lippai, D.; Catalano, D.; Mandrekar, P.; Dolganiuc, A.; Kurt-Jones, E.; et al. Deficiency in myeloid differentiation factor-2 and toll-like receptor 4 expression attenuates nonalcoholic steatohepatitis and fibrosis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G433–G441. [Google Scholar] [CrossRef]
- Yang, B.; Luo, W.; Wang, M.; Tang, Y.; Zhu, W.; Jin, L.; Wang, M.; Wang, Y.; Zhang, Y.; Zuo, W.; et al. Macrophage-specific MyD88 deletion and pharmacological inhibition prevents liver damage in non-alcoholic fatty liver disease via reducing inflammatory response. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166480. [Google Scholar] [CrossRef]
- Gong, J.; Li, J.; Dong, H.; Chen, G.; Qin, X.; Hu, M.; Yuan, F.; Fang, K.; Wang, D.; Jiang, S.; et al. Inhibitory effects of berberine on proinflammatory M1 macrophage polarization through interfering with the interaction between TLR4 and MyD88. BMC Complement. Altern. Med. 2019, 19, 314. [Google Scholar] [CrossRef]
- Xiang, P.; Chen, T.; Mou, Y.; Wu, H.; Xie, P.; Lu, G.; Gong, X.; Hu, Q.; Zhang, Y.; Ji, H. NZ suppresses TLR4/NF-kappaB signalings and NLRP3 inflammasome activation in LPS-induced RAW264.7 macrophages. Inflamm. Res. 2015, 64, 799–808. [Google Scholar] [CrossRef]
- Lu, H.; Wu, L.; Liu, L.; Ruan, Q.; Zhang, X.; Hong, W.; Wu, S.; Jin, G.; Bai, Y. Quercetin ameliorates kidney injury and fibrosis by modulating M1/M2 macrophage polarization. Biochem. Pharmacol. 2018, 154, 203–212. [Google Scholar] [CrossRef]
- Koperska, A.; Wesolek, A.; Moszak, M.; Szulinska, M. Berberine in Non-Alcoholic Fatty Liver Disease-A Review. Nutrients 2022, 14, 3459. [Google Scholar] [CrossRef] [PubMed]
- Nie, Q.; Li, M.; Huang, C.; Yuan, Y.; Liang, Q.; Ma, X.; Qiu, T.; Li, J. The clinical efficacy and safety of berberine in the treatment of non-alcoholic fatty liver disease: A meta-analysis and systematic review. J. Transl. Med. 2024, 22, 225. [Google Scholar] [CrossRef]
- Ren, S.; Ma, X.; Wang, R.; Liu, H.; Wei, Y.; Wei, S.; Jing, M.; Zhao, Y. Preclinical Evidence of Berberine on Non-Alcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis of Animal Studies. Front. Pharmacol. 2021, 12, 742465. [Google Scholar] [CrossRef]
- Sotiropoulou, M.; Katsaros, I.; Vailas, M.; Lidoriki, I.; Papatheodoridis, G.V.; Kostomitsopoulos, N.G.; Valsami, G.; Tsaroucha, A.; Schizas, D. Nonalcoholic fatty liver disease: The role of quercetin and its therapeutic implications. Saudi J. Gastroenterol. 2021, 27, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, T.; Heng, C.; Zhou, Y.; Jiang, Z.; Qian, X.; Du, L.; Mao, S.; Yin, X.; Lu, Q. Quercetin improves nonalcoholic fatty liver by ameliorating inflammation, oxidative stress, and lipid metabolism in db/db mice. Phytother. Res. 2019, 33, 3140–3152. [Google Scholar] [CrossRef]
- Qi, Q.R.; Yang, Z.M. Regulation and function of signal transducer and activator of transcription 3. World J. Biol. Chem. 2014, 5, 231–239. [Google Scholar] [CrossRef]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Gao, Y.; Zhang, Q.; Zhou, G.; Cao, F.; Yao, S. IL-4 Switches Microglia/macrophage M1/M2 Polarization and Alleviates Neurological Damage by Modulating the JAK1/STAT6 Pathway Following ICH. Neuroscience 2020, 437, 161–171. [Google Scholar] [CrossRef]
- Quero, L.; Tiaden, A.N.; Hanser, E.; Roux, J.; Laski, A.; Hall, J.; Kyburz, D. miR-221-3p Drives the Shift of M2-Macrophages to a Pro-Inflammatory Function by Suppressing JAK3/STAT3 Activation. Front. Immunol. 2019, 10, 3087. [Google Scholar] [CrossRef]
- Travis, M.A.; Sheppard, D. TGF-beta activation and function in immunity. Annu. Rev. Immunol. 2014, 32, 51–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Wang, X.; Wang, P.; Essandoh, K.; Cui, S.; Huang, W.; Mu, X.; Liu, Z.; Wang, Y.; et al. GDF3 Protects Mice against Sepsis-Induced Cardiac Dysfunction and Mortality by Suppression of Macrophage Pro-Inflammatory Phenotype. Cells 2020, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Jin, W.; Guo, Z.; Hao, J. Quercetin Alleviates Asthma-Induced Airway Inflammation and Remodeling through Downregulating Periostin via Blocking TGF-beta1/Smad Pathway. Pharmacology 2023, 108, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Wahli, W.; Michalik, L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 2012, 23, 351–363. [Google Scholar] [CrossRef]
- Delerive, P.; De Bosscher, K.; Vanden Berghe, W.; Fruchart, J.C.; Haegeman, G.; Staels, B. DNA binding-independent induction of IkappaBalpha gene transcription by PPARalpha. Mol. Endocrinol. 2002, 16, 1029–1039. [Google Scholar] [CrossRef]
- Hou, Y.; Moreau, F.; Chadee, K. PPARgamma is an E3 ligase that induces the degradation of NFkappaB/p65. Nat. Commun. 2012, 3, 1300. [Google Scholar] [CrossRef]
- Toobian, D.; Ghosh, P.; Katkar, G.D. Parsing the Role of PPARs in Macrophage Processes. Front. Immunol. 2021, 12, 783780. [Google Scholar] [CrossRef]
- Yu, L.; Gao, Y.; Aaron, N.; Qiang, L. A glimpse of the connection between PPARgamma and macrophage. Front. Pharmacol. 2023, 14, 1254317. [Google Scholar] [CrossRef] [PubMed]
- Chinetti-Gbaguidi, G.; Staels, B. PPARbeta in macrophages and atherosclerosis. Biochimie 2017, 136, 59–64. [Google Scholar] [CrossRef]
- Cheng, L.; Liu, D.; Gao, S. PPARA ameliorates sepsis-induced myocardial injury via promoting macrophage M2 polarization by interacting with DUSP1. Regen Ther. 2024, 26, 33–41. [Google Scholar] [CrossRef]
- Luo, W.; Xu, Q.; Wang, Q.; Wu, H.; Hua, J. Effect of modulation of PPAR-gamma activity on Kupffer cells M1/M2 polarization in the development of non-alcoholic fatty liver disease. Sci. Rep. 2017, 7, 44612. [Google Scholar] [CrossRef]
- Bouhlel, M.A.; Derudas, B.; Rigamonti, E.; Dievart, R.; Brozek, J.; Haulon, S.; Zawadzki, C.; Jude, B.; Torpier, G.; Marx, N.; et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007, 6, 137–143. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Red Eagle, A.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef]
- Wu, D.; Molofsky, A.B.; Liang, H.E.; Ricardo-Gonzalez, R.R.; Jouihan, H.A.; Bando, J.K.; Chawla, A.; Locksley, R.M. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science 2011, 332, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ma, C.; Gong, L.; Guo, Y.; Fu, K.; Zhang, Y.; Zhou, H.; Li, Y. Macrophage Polarization and Its Role in Liver Disease. Front. Immunol. 2021, 12, 803037. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Wang, S.; Guo, X.; Wang, J.; Zhang, Z.; Qiu, W.; Gao, X.; Chen, Z.; Xu, J.; Zhao, R.; et al. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-kappaB pathways. Oncogene 2020, 39, 428–442. [Google Scholar] [CrossRef]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef]
- Singla, R.D.; Wang, J.; Singla, D.K. Regulation of Notch 1 signaling in THP-1 cells enhances M2 macrophage differentiation. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H1634–H1642. [Google Scholar] [CrossRef] [PubMed]
- Byles, V.; Covarrubias, A.J.; Ben-Sahra, I.; Lamming, D.W.; Sabatini, D.M.; Manning, B.D.; Horng, T. The TSC-mTOR pathway regulates macrophage polarization. Nat. Commun. 2013, 4, 2834. [Google Scholar] [CrossRef]
- Yu, T.; Gao, M.; Yang, P.; Liu, D.; Wang, D.; Song, F.; Zhang, X.; Liu, Y. Insulin promotes macrophage phenotype transition through PI3K/Akt and PPAR-gamma signaling during diabetic wound healing. J. Cell. Physiol. 2019, 234, 4217–4231. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, T.; Jiang, L.; Gao, J.; Yu, D.; Ge, Y.; Lin, S. MCP-induced protein 1 attenuates sepsis-induced acute lung injury by modulating macrophage polarization via the JNK/c-Myc pathway. Int. Immunopharmacol. 2019, 75, 105741. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ni, M.; Zong, R.; Yu, M.; Sun, Y.; Li, J.; Chen, P.; Li, C. Targeting Notch1-YAP Circuit Reprograms Macrophage Polarization and Alleviates Acute Liver Injury in Mice. Cell. Mol. Gastroenterol. Hepatol. 2023, 15, 1085–1104. [Google Scholar] [CrossRef] [PubMed]
- Panwar, V.; Singh, A.; Bhatt, M.; Tonk, R.K.; Azizov, S.; Raza, A.S.; Sengupta, S.; Kumar, D.; Garg, M. Multifaceted role of mTOR (mammalian target of rapamycin) signaling pathway in human health and disease. Signal Transduct. Target. Ther. 2023, 8, 375. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; O’Brien, T.F.; Zhang, P.; Zhong, X.P. The role of tuberous sclerosis complex 1 in regulating innate immunity. J. Immunol. 2012, 188, 3658–3666. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, T.; Li, L.; Sun, L.; Hou, Y.; Hu, X.; Zhang, L.; Tian, H.; Zhao, Q.; Peng, J.; et al. TSC1 controls macrophage polarization to prevent inflammatory disease. Nat. Commun. 2014, 5, 4696. [Google Scholar] [CrossRef]
- Jiang, H.; Westerterp, M.; Wang, C.; Zhu, Y.; Ai, D. Macrophage mTORC1 disruption reduces inflammation and insulin resistance in obese mice. Diabetologia 2014, 57, 2393–2404. [Google Scholar] [CrossRef] [PubMed]
- Ai, D.; Jiang, H.; Westerterp, M.; Murphy, A.J.; Wang, M.; Ganda, A.; Abramowicz, S.; Welch, C.; Almazan, F.; Zhu, Y.; et al. Disruption of mammalian target of rapamycin complex 1 in macrophages decreases chemokine gene expression and atherosclerosis. Circ. Res. 2014, 114, 1576–1584. [Google Scholar] [CrossRef]
- Maina, V.; Sutti, S.; Locatelli, I.; Vidali, M.; Mombello, C.; Bozzola, C.; Albano, E. Bias in macrophage activation pattern influences non-alcoholic steatohepatitis (NASH) in mice. Clin. Sci. 2012, 122, 545–553. [Google Scholar] [CrossRef]
- Wan, J.; Benkdane, M.; Teixeira-Clerc, F.; Bonnafous, S.; Louvet, A.; Lafdil, F.; Pecker, F.; Tran, A.; Gual, P.; Mallat, A.; et al. M2 Kupffer cells promote M1 Kupffer cell apoptosis: A protective mechanism against alcoholic and nonalcoholic fatty liver disease. Hepatology 2014, 59, 130–142. [Google Scholar] [CrossRef]
- Papackova, Z.; Palenickova, E.; Dankova, H.; Zdychova, J.; Skop, V.; Kazdova, L.; Cahova, M. Kupffer cells ameliorate hepatic insulin resistance induced by high-fat diet rich in monounsaturated fatty acids: The evidence for the involvement of alternatively activated macrophages. Nutr. Metab. 2012, 9, 22. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Red Eagle, A.; Vats, D.; Morel, C.R.; Goforth, M.H.; Subramanian, V.; Mukundan, L.; Ferrante, A.W.; Chawla, A. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008, 7, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Rolny, C.; Mazzone, M.; Tugues, S.; Laoui, D.; Johansson, I.; Coulon, C.; Squadrito, M.L.; Segura, I.; Li, X.; Knevels, E.; et al. HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF. Cancer Cell 2011, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Bartneck, M.; Fech, V.; Ehling, J.; Govaere, O.; Warzecha, K.T.; Hittatiya, K.; Vucur, M.; Gautheron, J.; Luedde, T.; Trautwein, C.; et al. Histidine-rich glycoprotein promotes macrophage activation and inflammation in chronic liver disease. Hepatology 2016, 63, 1310–1324. [Google Scholar] [CrossRef]
- Rensen, S.S.; Slaats, Y.; Nijhuis, J.; Jans, A.; Bieghs, V.; Driessen, A.; Malle, E.; Greve, J.W.; Buurman, W.A. Increased hepatic myeloperoxidase activity in obese subjects with nonalcoholic steatohepatitis. Am. J. Pathol. 2009, 175, 1473–1482. [Google Scholar] [CrossRef]
- Hart, K.M.; Fabre, T.; Sciurba, J.C.; Gieseck, R.L., 3rd; Borthwick, L.A.; Vannella, K.M.; Acciani, T.H.; de Queiroz Prado, R.; Thompson, R.W.; White, S.; et al. Type 2 immunity is protective in metabolic disease but exacerbates NAFLD collaboratively with TGF-beta. Sci. Transl. Med. 2017, 9, eaal3694. [Google Scholar] [CrossRef]
- Nakayama, T.; Hirahara, K.; Onodera, A.; Endo, Y.; Hosokawa, H.; Shinoda, K.; Tumes, D.J.; Okamoto, Y. Th2 Cells in Health and Disease. Annu. Rev. Immunol. 2017, 35, 53–84. [Google Scholar] [CrossRef]
- Svendsen, P.; Graversen, J.H.; Etzerodt, A.; Hager, H.; Roge, R.; Gronbaek, H.; Christensen, E.I.; Moller, H.J.; Vilstrup, H.; Moestrup, S.K. Antibody-Directed Glucocorticoid Targeting to CD163 in M2-type Macrophages Attenuates Fructose-Induced Liver Inflammatory Changes. Mol. Ther. Methods Clin. Dev. 2017, 4, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Guillot, A.; Tacke, F. Liver Macrophages: Old Dogmas and New Insights. Hepatol. Commun. 2019, 3, 730–743. [Google Scholar] [CrossRef]
- Daemen, S.; Gainullina, A.; Kalugotla, G.; He, L.; Chan, M.M.; Beals, J.W.; Liss, K.H.; Klein, S.; Feldstein, A.E.; Finck, B.N.; et al. Dynamic Shifts in the Composition of Resident and Recruited Macrophages Influence Tissue Remodeling in NASH. Cell Rep. 2021, 34, 108626. [Google Scholar] [CrossRef]
- Bonnardel, J.; T’Jonck, W.; Gaublomme, D.; Browaeys, R.; Scott, C.L.; Martens, L.; Vanneste, B.; De Prijck, S.; Nedospasov, S.A.; Kremer, A.; et al. Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 2019, 51, 638–654.e639. [Google Scholar] [CrossRef]
- Mulder, K.; Patel, A.A.; Kong, W.T.; Piot, C.; Halitzki, E.; Dunsmore, G.; Khalilnezhad, S.; Irac, S.E.; Dubuisson, A.; Chevrier, M.; et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 2021, 54, 1883–1900.e5. [Google Scholar] [CrossRef] [PubMed]
- Khantakova, D.; Brioschi, S.; Molgora, M. Exploring the Impact of TREM2 in Tumor-Associated Macrophages. Vaccines 2022, 10, 943. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, M.; Guo, H.; Hou, J.; Zhang, Y.; Li, M.; Wu, X.; Chen, X.; Wang, L. Integrated Analysis Highlights the Immunosuppressive Role of TREM2(+) Macrophages in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 848367. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M. The biology of TREM receptors. Nat. Rev. Immunol. 2023, 23, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Labiano, I.; Agirre-Lizaso, A.; Olaizola, P.; Echebarria, A.; Huici-Izagirre, M.; Olaizola, I.; Esparza-Baquer, A.; Sharif, O.; Hijona, E.; Milkiewicz, P.; et al. TREM-2 plays a protective role in cholestasis by acting as a negative regulator of inflammation. J. Hepatol. 2022, 77, 991–1004. [Google Scholar] [CrossRef]
- Hendrikx, T.; Porsch, F.; Kiss, M.G.; Rajcic, D.; Papac-Milicevic, N.; Hoebinger, C.; Goederle, L.; Hladik, A.; Shaw, L.E.; Horstmann, H.; et al. Soluble TREM2 levels reflect the recruitment and expansion of TREM2(+) macrophages that localize to fibrotic areas and limit NASH. J. Hepatol. 2022, 77, 1373–1385. [Google Scholar] [CrossRef]
- Coelho, I.; Duarte, N.; Barros, A.; Macedo, M.P.; Penha-Goncalves, C. Trem-2 Promotes Emergence of Restorative Macrophages and Endothelial Cells during Recovery from Hepatic Tissue Damage. Front. Immunol. 2020, 11, 616044. [Google Scholar] [CrossRef]
- Krenkel, O.; Hundertmark, J.; Abdallah, A.T.; Kohlhepp, M.; Puengel, T.; Roth, T.; Branco, D.P.P.; Mossanen, J.C.; Luedde, T.; Trautwein, C.; et al. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut 2020, 69, 551–563. [Google Scholar] [CrossRef]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Toth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017, 542, 352–356. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Guillot, A.; Tacke, F. Spatial dimension of macrophage heterogeneity in liver diseases. eGastroenterology 2023, 1, e000003. [Google Scholar] [CrossRef]
- Guilliams, M.; Bonnardel, J.; Haest, B.; Vanderborght, B.; Wagner, C.; Remmerie, A.; Bujko, A.; Martens, L.; Thone, T.; Browaeys, R.; et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 2022, 185, 379–396.e338. [Google Scholar] [CrossRef] [PubMed]
- Gola, A.; Dorrington, M.G.; Speranza, E.; Sala, C.; Shih, R.M.; Radtke, A.J.; Wong, H.S.; Baptista, A.P.; Hernandez, J.M.; Castellani, G.; et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 2021, 589, 131–136. [Google Scholar] [CrossRef]
- Andrews, T.S.; Atif, J.; Liu, J.C.; Perciani, C.T.; Ma, X.Z.; Thoeni, C.; Slyper, M.; Eraslan, G.; Segerstolpe, A.; Manuel, J.; et al. Single-Cell, Single-Nucleus, and Spatial RNA Sequencing of the Human Liver Identifies Cholangiocyte and Mesenchymal Heterogeneity. Hepatol. Commun. 2022, 6, 821–840. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.G.; Sekine, K.; Gerber, T.; Loeffler-Wirth, H.; Binder, H.; Gac, M.; Kanton, S.; Kageyama, J.; Damm, G.; Seehofer, D.; et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017, 546, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, M.; Diamanti, K.; Pan, G.; Spalinskas, R.; Kumar, C.; Deshmukh, A.S.; Mann, M.; Sahlen, P.; Komorowski, J.; Wadelius, C. A Multi-Omics Approach to Liver Diseases: Integration of Single Nuclei Transcriptomics with Proteomics and HiCap Bulk Data in Human Liver. OMICS 2020, 24, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Dudek, M.; Pfister, D.; Donakonda, S.; Filpe, P.; Schneider, A.; Laschinger, M.; Hartmann, D.; Huser, N.; Meiser, P.; Bayerl, F.; et al. Auto-aggressive CXCR6(+) CD8 T cells cause liver immune pathology in NASH. Nature 2021, 592, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Yang, Y.; Li, P.; Zeng, Z.; Hu, W.; Zhe, R.; Liu, X.; Tang, D.; Ou, M.; Dai, Y. Integrating Spatial Transcriptomics and Single-Cell RNA-seq Reveals the Gene Expression Profling of the Human Embryonic Liver. Front. Cell Dev. Biol. 2021, 9, 652408. [Google Scholar] [CrossRef]
- Payen, V.L.; Lavergne, A.; Alevra Sarika, N.; Colonval, M.; Karim, L.; Deckers, M.; Najimi, M.; Coppieters, W.; Charloteaux, B.; Sokal, E.M.; et al. Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep. 2021, 3, 100278. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Keogh, A.; Waldt, A.; Cuttat, R.; Neri, M.; Zhu, S.; Schuierer, S.; Ruchti, A.; Crochemore, C.; Knehr, J.; et al. Single-cell and bulk transcriptomics of the liver reveals potential targets of NASH with fibrosis. Sci. Rep. 2021, 11, 19396. [Google Scholar] [CrossRef]
- Filliol, A.; Saito, Y.; Nair, A.; Dapito, D.H.; Yu, L.X.; Ravichandra, A.; Bhattacharjee, S.; Affo, S.; Fujiwara, N.; Su, H.; et al. Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis. Nature 2022, 610, 356–365. [Google Scholar] [CrossRef]
- Kotsiliti, E.; Leone, V.; Schuehle, S.; Govaere, O.; Li, H.; Wolf, M.J.; Horvatic, H.; Bierwirth, S.; Hundertmark, J.; Inverso, D.; et al. Intestinal B cells license metabolic T-cell activation in NASH microbiota/antigen-independently and contribute to fibrosis by IgA-FcR signalling. J. Hepatol. 2023, 79, 296–313. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, K.; Pickholz, E.; Dobie, R.; Matchett, K.P.; Henderson, N.C.; Carrico, C.; Driver, I.; Borch Jensen, M.; Chen, L.; et al. An autocrine signaling circuit in hepatic stellate cells underlies advanced fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med. 2023, 15, eadd3949. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Batmanov, K.; Hu, W.; Zhu, K.; Tom, A.Y.; Guan, D.; Jiang, C.; Cheng, L.; McCright, S.J.; Yang, E.C.; et al. Hepatocytes demarcated by EphB2 contribute to the progression of nonalcoholic steatohepatitis. Sci. Transl. Med. 2023, 15, eadc9653. [Google Scholar] [CrossRef] [PubMed]
- Guillot, A.; Winkler, M.; Silva Afonso, M.; Aggarwal, A.; Lopez, D.; Berger, H.; Kohlhepp, M.S.; Liu, H.; Ozdirik, B.; Eschrich, J.; et al. Mapping the hepatic immune landscape identifies monocytic macrophages as key drivers of steatohepatitis and cholangiopathy progression. Hepatology 2023, 78, 150–166. [Google Scholar] [CrossRef]
- Ait Ahmed, Y.; Lafdil, F.; Tacke, F. Ambiguous Pathogenic Roles of Macrophages in Alcohol-Associated Liver Diseases. Hepat. Med. 2023, 15, 113–127. [Google Scholar] [CrossRef]
- Peiseler, M.; Araujo David, B.; Zindel, J.; Surewaard, B.G.J.; Lee, W.Y.; Heymann, F.; Nusse, Y.; Castanheira, F.V.S.; Shim, R.; Guillot, A.; et al. Kupffer cell-like syncytia replenish resident macrophage function in the fibrotic liver. Science 2023, 381, eabq5202. [Google Scholar] [CrossRef]
- Stienstra, R.; Saudale, F.; Duval, C.; Keshtkar, S.; Groener, J.E.; van Rooijen, N.; Staels, B.; Kersten, S.; Muller, M. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology 2010, 51, 511–522. [Google Scholar] [CrossRef]
- Huang, W.; Metlakunta, A.; Dedousis, N.; Zhang, P.; Sipula, I.; Dube, J.J.; Scott, D.K.; O’Doherty, R.M. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 2010, 59, 347–357. [Google Scholar] [CrossRef]
- Kudo, H.; Takahara, T.; Yata, Y.; Kawai, K.; Zhang, W.; Sugiyama, T. Lipopolysaccharide triggered TNF-alpha-induced hepatocyte apoptosis in a murine non-alcoholic steatohepatitis model. J. Hepatol. 2009, 51, 168–175. [Google Scholar] [CrossRef]
- Ratziu, V.; Sanyal, A.; Harrison, S.A.; Wong, V.W.; Francque, S.; Goodman, Z.; Aithal, G.P.; Kowdley, K.V.; Seyedkazemi, S.; Fischer, L.; et al. Cenicriviroc Treatment for Adults with Nonalcoholic Steatohepatitis and Fibrosis: Final Analysis of the Phase 2b CENTAUR Study. Hepatology 2020, 72, 892–905. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Marri, S.R.; Chalasani, N.; Kohli, R.; Aronstein, W.; Thompson, G.A.; Irish, W.; Miles, M.V.; Xanthakos, S.A.; Lawitz, E.; et al. Randomised clinical study: GR-MD-02, a galectin-3 inhibitor, vs. placebo in patients having non-alcoholic steatohepatitis with advanced fibrosis. Aliment. Pharmacol. Ther. 2016, 44, 1183–1198. [Google Scholar] [CrossRef]
- Friedman, S.L.; Ratziu, V.; Harrison, S.A.; Abdelmalek, M.F.; Aithal, G.P.; Caballeria, J.; Francque, S.; Farrell, G.; Kowdley, K.V.; Craxi, A.; et al. A randomized, placebo-controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology 2018, 67, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.K.; Yang, R.Y.; Pan, Z.; Yu, L.; Salomon, D.R.; Fung-Leung, W.P.; Liu, F.T. Targeted disruption of the galectin-3 gene results in attenuated peritoneal inflammatory responses. Am. J. Pathol. 2000, 156, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Hsu, D.K.; Zuberi, R.I.; Kuwabara, I.; Chi, E.Y.; Henderson, W.R., Jr. Expression and function of galectin-3, a beta-galactoside-binding lectin, in human monocytes and macrophages. Am. J. Pathol. 1995, 147, 1016–1028. [Google Scholar]
- Hughes, R.C. Galectins as modulators of cell adhesion. Biochimie 2001, 83, 667–676. [Google Scholar] [CrossRef]
- Di Lella, S.; Sundblad, V.; Cerliani, J.P.; Guardia, C.M.; Estrin, D.A.; Vasta, G.R.; Rabinovich, G.A. When galectins recognize glycans: From biochemistry to physiology and back again. Biochemistry 2011, 50, 7842–7857. [Google Scholar] [CrossRef]
- Iacobini, C.; Menini, S.; Ricci, C.; Blasetti Fantauzzi, C.; Scipioni, A.; Salvi, L.; Cordone, S.; Delucchi, F.; Serino, M.; Federici, M.; et al. Galectin-3 ablation protects mice from diet-induced NASH: A major scavenging role for galectin-3 in liver. J. Hepatol. 2011, 54, 975–983. [Google Scholar] [CrossRef]
- Jeftic, I.; Jovicic, N.; Pantic, J.; Arsenijevic, N.; Lukic, M.L.; Pejnovic, N. Galectin-3 Ablation Enhances Liver Steatosis, but Attenuates Inflammation and IL-33-Dependent Fibrosis in Obesogenic Mouse Model of Nonalcoholic Steatohepatitis. Mol. Med. 2015, 21, 453–465. [Google Scholar] [CrossRef]
- Traber, P.G.; Zomer, E. Therapy of experimental NASH and fibrosis with galectin inhibitors. PLoS ONE 2013, 8, e83481. [Google Scholar] [CrossRef]
- Chalasani, N.; Abdelmalek, M.F.; Garcia-Tsao, G.; Vuppalanchi, R.; Alkhouri, N.; Rinella, M.; Noureddin, M.; Pyko, M.; Shiffman, M.; Sanyal, A.; et al. Effects of Belapectin, an Inhibitor of Galectin-3, in Patients with Nonalcoholic Steatohepatitis with Cirrhosis and Portal Hypertension. Gastroenterology 2020, 158, 1334–1345.e1335. [Google Scholar] [CrossRef]
- Noureddin, M. MASH clinical trials and drugs pipeline: An impending tsunami. Hepatology 2024. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, P.; Ruan, X.; Ma, Y.; Kawai, K.; Suemizu, H.; Cao, H. Comparative Transcriptomics Analyses in Livers of Mice, Humans, and Humanized Mice Define Human-Specific Gene Networks. Cells 2020, 9, 2566. [Google Scholar] [CrossRef] [PubMed]
- Teufel, A.; Itzel, T.; Erhart, W.; Brosch, M.; Wang, X.Y.; Kim, Y.O.; von Schonfels, W.; Herrmann, A.; Bruckner, S.; Stickel, F.; et al. Comparison of Gene Expression Patterns between Mouse Models of Nonalcoholic Fatty Liver Disease and Liver Tissues from Patients. Gastroenterology 2016, 151, 513–525.e510. [Google Scholar] [CrossRef] [PubMed]
- Saldarriaga, O.A.; Wanninger, T.G.; Arroyave, E.; Gosnell, J.; Krishnan, S.; Oneka, M.; Bao, D.; Millian, D.E.; Kueht, M.L.; Moghe, A.; et al. Heterogeneity in intrahepatic macrophage populations and druggable target expression in patients with steatotic liver disease-related fibrosis. JHEP Rep. 2024, 6, 100958. [Google Scholar] [CrossRef]
- Guillot, A.; Tacke, F. Liver macrophages revisited: The expanding universe of versatile responses in a spatiotemporal context. Hepatol. Commun. 2024, 8, e0491. [Google Scholar] [CrossRef]
| Method | Tissues | Database Accession | Publications |
|---|---|---|---|
| scRNAseq | Fetal livers (10.5- and 17.5-week gestation) and healthy liver tissues from partial hepatectomy of patients with primary or secondary liver tumors or benign liver diseases (n = 3) | GSE81252 and GSE96981 | [185] |
| scRNAseq | Livers from donors for liver transplantation (n = 5) | GSE115469 | [64] |
| scRNAseq | Non-diseased liver tissues from patients who underwent liver resections for colorectal cancer metastasis or cholangiocarcinoma (n = 6) | GSE124395 | [66] |
| scRNAseq | Non-parenchymal cells from healthy livers (n = 5) and cirrhotic livers (two MASLD, two ALD, one PBC) | GSE136103 | [68] |
| snRNAseq | Tumor-free liver tissue from a partial hepatectomy of a patient with colon cancer and hepatic metastasis (n = 1) | EBI BioStudies S-BSST324 | [186] |
| scRNAseq | Liver CD45+ immune cells from healthy livers (n = 3) | GSE125188 | [65] |
| scRNAseq | Liver CD45+ immune cells from steatosis (n = 4) and MASH livers (n = 3) | GSE159977 | [187] |
| scRNAseq; Spatial transcriptomics | Fetal livers (8- and 17-week gestation) | GSE167096 | [188] |
| scRNAseq | Liver tissues from donors (n = 2) | GSE158723 | [189] |
| scRNAseq | Healthy livers from donors (n = 6) | EBI BioStudies E-MTAB-10553 | [190] |
| scRNAseq; snRNAseq; Spatial transcriptomics | Healthy livers from donors for liver transplantation (n = 4) | GSE185477 | [184] |
| snRNAseq | Healthy livers (n = 2), MASLD cirrhotic livers (n = 2), HCC (n = 2), and adjacent cirrhotic livers (n = 2) | GSE174748 and GSE212047 | [191] |
| scRNAseq; snRNAseq; CITE-seq | Healthy (n = 14), >10% steatosis with no fibrosis (n = 5) | GSE192742 | [182] |
| scRNAseq | Healthy (n = 1) and cirrhotic MASH liver (n = 1) | GSE190487 and GSM5724573 | [192] |
| snRNAseq | Normal livers (non-tumor tissue from liver metastasis resections) (n = 3) and MASH livers (n = 9) | GSE212837 | [193] |
| snRNAseq | Healthy livers (n = 3) and MASH livers (n = 3) | GSE189600 | [194] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahamed, F.; Eppler, N.; Jones, E.; Zhang, Y. Understanding Macrophage Complexity in Metabolic Dysfunction-Associated Steatotic Liver Disease: Transitioning from the M1/M2 Paradigm to Spatial Dynamics. Livers 2024, 4, 455-478. https://doi.org/10.3390/livers4030033
Ahamed F, Eppler N, Jones E, Zhang Y. Understanding Macrophage Complexity in Metabolic Dysfunction-Associated Steatotic Liver Disease: Transitioning from the M1/M2 Paradigm to Spatial Dynamics. Livers. 2024; 4(3):455-478. https://doi.org/10.3390/livers4030033
Chicago/Turabian StyleAhamed, Forkan, Natalie Eppler, Elizabeth Jones, and Yuxia Zhang. 2024. "Understanding Macrophage Complexity in Metabolic Dysfunction-Associated Steatotic Liver Disease: Transitioning from the M1/M2 Paradigm to Spatial Dynamics" Livers 4, no. 3: 455-478. https://doi.org/10.3390/livers4030033
APA StyleAhamed, F., Eppler, N., Jones, E., & Zhang, Y. (2024). Understanding Macrophage Complexity in Metabolic Dysfunction-Associated Steatotic Liver Disease: Transitioning from the M1/M2 Paradigm to Spatial Dynamics. Livers, 4(3), 455-478. https://doi.org/10.3390/livers4030033






