Redrawing the Map to Novel DILI Biomarkers in Circulation: Where Are We, Where Should We Go, and How Can We Get There?
Abstract
:1. Introduction
2. Points of Departure
2.1. Where Are We?
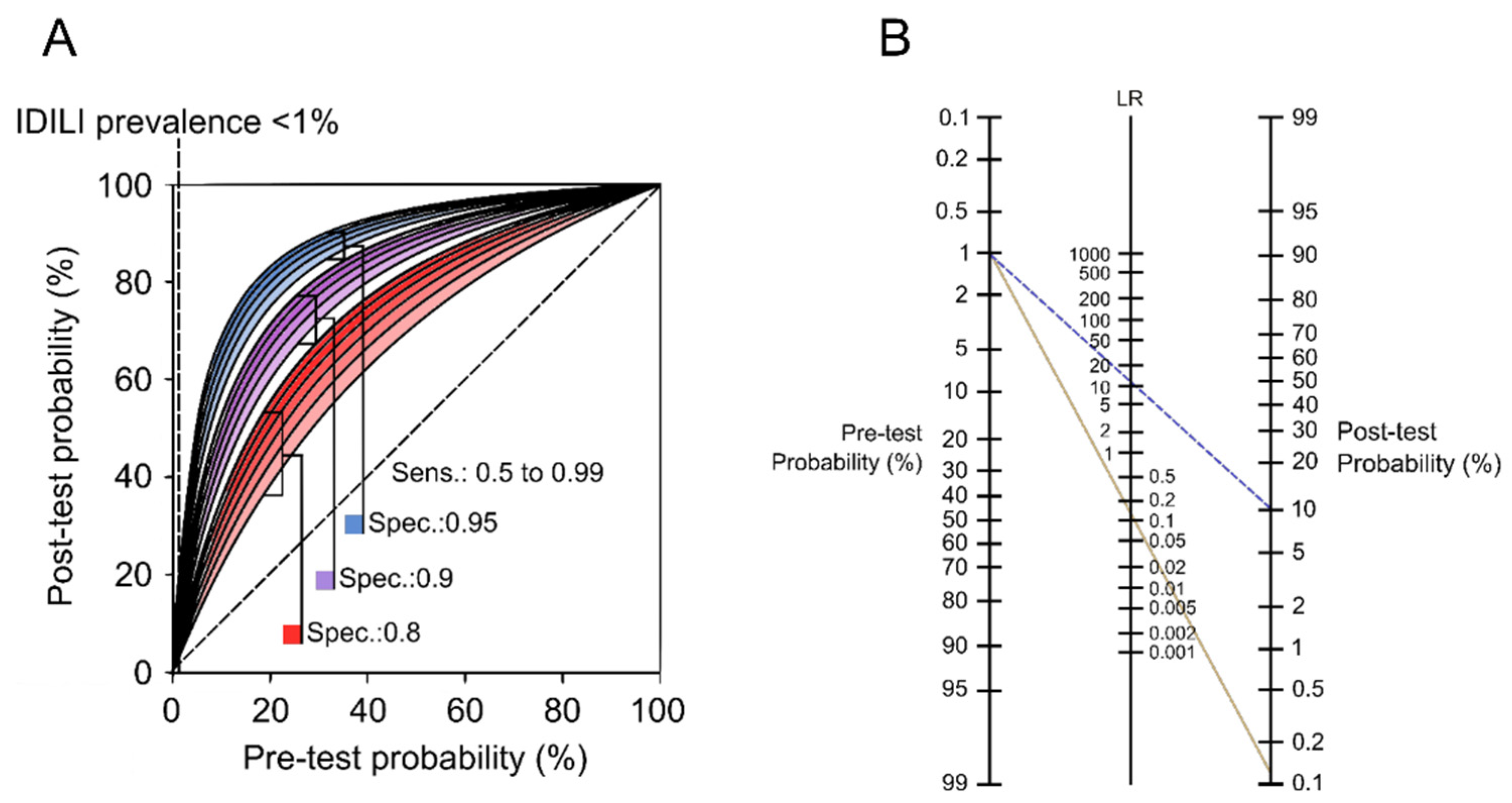
2.2. How Did We Get Here?
2.3. How Should We Move Forward?
3. DILI Destinations
3.1. Biomarkers for Diagnosis
3.2. Biomarkers for Prediction
3.3. Biomarkers for Prognosis
3.4. Biomarkers for Hepatotoxic Liability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, W.M. Acute Liver Failure. Semin. Respir. Crit. Care Med. 2012, 33, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Mezzapelle, R.; Venereau, E.; Bianchi, M.E. Stress and Alarmins. Report from the 9th iD&EAs meeting. Cell Death Dis. 2019, 10, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Craig, D.G.N.; Lee, P.; Pryde, E.A.; Masterton, G.S.; Hayes, P.C.; Simpson, K.J. Circulating apoptotic and necrotic cell death markers in patients with acute liver injury. Liver Int. 2011, 31, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Schomaker, S.; Warner, R.; Bock, J.; Johnson, K.; Potter, D.; Van Winkle, J.; Aubrecht, J. Assessment of Emerging Biomarkers of Liver Injury in Human Subjects. Toxicol. Sci. 2013, 132, 276–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGill, M.; Sharpe, M.R.; Williams, C.D.; Taha, M.; Curry, S.C.; Jaeschke, H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J. Clin. Investig. 2012, 122, 1574–1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGill, M.R.; Staggs, V.S.; Sharpe, M.R.; Lee, W.M.; Jaeschke, H. Acute Liver Failure Study Group Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology 2014, 60, 1336–1345. [Google Scholar] [CrossRef] [Green Version]
- Weemhoff, J.L.; Woolbright, B.L.; Jenkins, R.E.; McGill, M.; Sharpe, M.R.; Olson, J.C.; Antoine, D.J.; Curry, S.C.; Jaeschke, H. Plasma biomarkers to study mechanisms of liver injury in patients with hypoxic hepatitis. Liver Int. 2017, 37, 377–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dear, J.W.; Clarke, J.I.; Francis, B.; Allen, L.; Wraight, J.; Shen, J.; Dargan, P.; Wood, D.; Cooper, J.; Thomas, S.H.L.; et al. Risk stratification after paracetamol overdose using mechanistic biomarkers: Results from two prospective cohort studies. Lancet Gastroenterol. Hepatol. 2018, 3, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Church, R.J.; Kullak-Ublick, G.A.; Aubrecht, J.; Bonkovsky, H.L.; Chalasani, N.; Fontana, R.J.; Goepfert, J.C.; Hackman, F.; King, N.M.P.; Kirby, S.; et al. Candidate biomarkers for the diagnosis and prognosis of drug-induced liver injury: An international collaborative effort. Hepatology 2019, 69, 760–773. [Google Scholar] [CrossRef]
- McGill, M.; Jaeschke, H. Biomarkers of drug-induced liver injury: Progress and utility in research, medicine, and regulation. Expert Rev. Mol. Diagn. 2018, 18, 797–807. [Google Scholar] [CrossRef]
- Andrade, R.J.; Chalasani, N.; Björnsson, E.S.; Suzuki, A.; Kullak-Ublick, G.A.; Watkins, P.B.; Devarbhavi, H.; Merz, M.; Lucena, M.I.; Kaplowitz, N.; et al. Drug-induced liver injury. Nat. Rev. Dis. Prim. 2019, 5, 1–22. [Google Scholar] [CrossRef] [Green Version]
- McGill, M.; Li, F.; Sharpe, M.R.; Williams, C.D.; Curry, S.C.; Ma, X.; Jaeschke, H. Circulating acylcarnitines as biomarkers of mitochondrial dysfunction after acetaminophen overdose in mice and humans. Arch. Toxicol. 2013, 88, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, S.; Yan, K.; Pence, L.; Simpson, P.M.; Gill, P.; Letzig, L.G.; Beger, R.D.; E Sullivan, J.; Kearns, G.L.; Reed, M.D.; et al. Targeted liquid chromatography–mass spectrometry analysis of serum acylcarnitines in acetaminophen toxicity in children. Biomark. Med. 2014, 8, 147–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weerasinghe, S.V.W.; Jang, Y.-J.; Fontana, R.J.; Omary, M.B. Carbamoyl phosphate synthetase-1 is a rapid turnover biomarker in mouse and human acute liver injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G355–G364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGill, M.R.; Jaeschke, H. Biomarkers of mitotoxicity after acute liver injury: Further insights into the interpretation of glutamate dehydrogenase. J. Clin. Transl. Res. 2021, 7, 61–65. [Google Scholar] [CrossRef]
- Schmidt, L.E.; Dalhoff, K. Alpha-fetoprotein is a predictor of outcome in acetaminophen-induced liver injury. Hepatology 2005, 41, 26–31. [Google Scholar] [CrossRef]
- Schomaker, S.; Potter, D.; Warner, R.; Larkindale, J.; King, N.; Porter, A.C.; Owens, J.; Tomlinson, L.; Sauer, J.-M.; Johnson, K.; et al. Serum glutamate dehydrogenase activity enables early detection of liver injury in subjects with underlying muscle impairments. PLoS ONE 2020, 15, e0229753. [Google Scholar] [CrossRef]
- Leers, M.P.G.; Bergman, T.; Tribbick, G.; Persson, B.; Ramaekers, F.C.S.; Nap, M.; Schutte, B. Immunocytochemical detection and mapping of a cytokeratin 18 neo-epitope exposed during early apoptosis. J. Pathol. 1999, 187, 567–572. [Google Scholar] [CrossRef]
- Ueno, T.; Toi, M.; Bivén, K.; Bando, H.; Ogawa, T.; Linder, S. Measurement of an apoptotic product in the sera of breast cancer patients. Eur. J. Cancer 2003, 39, 769–774. [Google Scholar] [CrossRef]
- Linder, S.; Havelka, A.M.; Ueno, T.; Shoshan, M.C. Determining tumor apoptosis and necrosis in patient serum using cytokeratin 18 as a biomarker. Cancer Lett. 2004, 214, 1–9. [Google Scholar] [CrossRef]
- Bantel, H.; Lügering, A.; Heidemann, J.; Volkmann, X.; Poremba, C.; Strassburg, C.P.; Manns, M.P.; Schulze-Osthoff, K. Detection of apoptotic caspase activation in sera from patients with chronic HCV infection is associated with fibrotic liver injury. Hepatology 2004, 40, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Papatheodoridis, G.V.; Hadziyannis, E.; Tsochatzis, E.; Chrysanthos, N.; Georgiou, A.; Kafiri, G.; Manolakopoulos, S.; Tiniakos, D.G.; Giannousis, I.; Manesis, E.K.; et al. Serum apoptotic caspase activity as a marker of severity in HBeAg-negative chronic hepatitis B virus infection. Gut 2007, 57, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a Late Mediator of Endotoxin Lethality in Mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Rovere-Querini, P.; Capobianco, A.; Scaffidi, P.; Valentinis, B.; Catalanotti, F.; Giazzon, M.; Dumitriu, I.E.; Müller-Knapp, S.; Iannacone, M.; Traversari, C.; et al. HMGB1 is an endogenous immune adjuvant released by necrotic cells. EMBO Rep. 2004, 5, 825–830. [Google Scholar] [CrossRef] [PubMed]
- McGill, M.R. The past and present of serum aminotransferases and the future of liver injury biomarkers. EXCLI J. 2016, 15, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Karmen, A.; Wróblewski, F.; LaDue, J.S. Transaminase Activity in Human Blood. J. Clin. Investig. 1955, 34, 126–133. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef] [Green Version]
- Willeit, P.; Skroblin, P.; Moschen, A.R.; Yin, X.; Kaudewitz, D.; Zampetaki, A.; Barwari, T.; Whitehead, M.; Ramírez, C.M.; Goedeke, L.; et al. Circulating MicroRNA-122 Is Associated with the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes 2016, 66, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, F.G.; Tolkach, Y.; Deng, M.; Schmidt, D.; Perner, S.; Kristiansen, G.; Müller, S.C.; Ellinger, J. Serum miR-122-5p and miR-206 expression: Non-invasive prognostic biomarkers for renal cell carcinoma. Clin. Epigenetics 2018, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Umbaugh, D.S.; Jaeschke, H. Biomarkers of drug-induced liver injury: A mechanistic perspective through acetaminophen hepatotoxicity. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 363–375. [Google Scholar] [CrossRef]
- Senior, J.R. Alanine Aminotransferase: A Clinical and Regulatory Tool for Detecting Liver Injury–Past, Present, and Future. Clin. Pharmacol. Ther. 2012, 92, 332–339. [Google Scholar] [CrossRef]
- Alfirevic, A.; Pirmohamed, M. Predictive Genetic Testing for Drug-Induced Liver Injury: Considerations of Clinical Utility. Clin. Pharmacol. Ther. 2012, 92, 376–380. [Google Scholar] [CrossRef] [PubMed]
- McGill, M.R.; Jaeschke, H. Biomarkers of drug-induced liver injury. Adv. Pharmacol. 2019, 85, 221–239. [Google Scholar] [CrossRef]
- Roberts, D.W.; Lee, W.M.; Hinson, J.A.; Bai, S.; Swearingen, C.J.; Stravitz, R.T.; Reuben, A.; Letzig, L.; Simpson, P.M.; Rule, J.; et al. An Immunoassay to Rapidly Measure Acetaminophen Protein Adducts Accurately Identifies Patients with Acute Liver Injury or Failure. Clin. Gastroenterol. Hepatol. 2017, 15, 555–562.e3. [Google Scholar] [CrossRef] [Green Version]
- Urban, T.J.; Aithal, G.; Daly, A. Genetic Basis of Drug-Induced Liver Injury: Present and Future. Semin. Liver Dis. 2014, 34, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, S.; McGuirk, S.; Powell, G.; Cutrell, A.; Naderer, O.; Spreen, B.; Lafon, S.; Pearce, G.; Steel, H. Hypersensitivity reactions during therapy with the nucleoside reverse transcriptase inhibitor abacavir. Clin. Ther. 2001, 23, 1603–1614. [Google Scholar] [CrossRef]
- Reuben, A.; Tillman, H.; Fontana, R.J.; Davern, T.; McGuire, B.; Stravitz, R.T.; Durkalski, V.; Larson, A.M.; Liou, I.; Fix, O.; et al. Outcomes in Adults with Acute Liver Failure Between 1998 and 2013. Ann. Intern. Med. 2016, 164, 724–732. [Google Scholar] [CrossRef]
- Thanapirom, K.; Treeprasertsuk, S.; Soonthornworasiri, N.; Poovorawan, K.; Chaiteerakij, R.; Komolmit, P.; Phaosawasdi, K.; Pinzani, M. The incidence, etiologies, outcomes, and predictors of mortality of acute liver failure in Thailand: A population-base study. BMC Gastroenterol. 2019, 19, 18. [Google Scholar] [CrossRef]
- Warrillow, S.; Bailey, M.; Pilcher, D.; Kazemi, A.; McArthur, C.; Young, P.; Bellomo, R. Characteristics and outcomes of patients with acute liver failure admitted to Australian and New Zealand intensive care units. Intern. Med. J. 2019, 49, 874–885. [Google Scholar] [CrossRef]
- De Clercq, P.; Geerts, A.; Van Vlierberghe, H.; Verhelst, X. The utility of biomarkers in prognosis assessment of patients with acute liver failure. Hepatol. Res. 2021, 51, 750–757. [Google Scholar] [CrossRef]
- Xiao, L.-L.; Zhang, F.; Zhao, Y.-L.; Zhang, L.-J.; Xie, Z.-Y.; Huang, K.-Z.; Ouyang, X.-X.; Wu, X.-X.; Xu, X.-W.; Li, L.-J. Using advanced oxidation protein products and ischae-mia-modified albumin to monitor oxidative stress levels in patients with drug-induced liver injury. Sci. Rep. 2020, 10, 18128. [Google Scholar] [CrossRef]
- Olson, H.; Betton, G.; Robinson, D.; Thomas, K.; Monro, A.; Kolaja, G.; Lilly, P.; Sanders, J.; Sipes, G.; Bracken, W.; et al. Concordance of the Toxicity of Pharmaceuticals in Humans and in Animals. Regul. Toxicol. Pharmacol. 2000, 32, 56–67. [Google Scholar] [CrossRef]
- FDA; CDER. Guidance for Industry Drug-Induced Liver Injury: Premarketing Clinical Evaluation; FDA; CDER: Silver Spring, MD, USA, 2009. [Google Scholar]
- Watkins, P.B.; Kaplowitz, N.; Slattery, J.T.; Colonese, C.R.; Colucci, S.V.; Stewart, P.W.; Harris, S.C. Aminotransferase Elevations in Healthy Adults Receiving 4 Grams of Acetaminophen Daily. JAMA 2006, 296, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heard, K.J.; Green, J.L.; Dart, R.C. Serum Alanine Aminotransferase Elevation During 10 Days of Acetaminophen Use in Nondrinkers. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2010, 30, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Heard, K.; Green, J.L.; Anderson, V.; Bucher-Bartelson, B.; Dart, R.C. A randomized, placebo-controlled trial to determine the course of aminotransferase elevation during prolonged acetaminophen administration. BMC Pharmacol. Toxicol. 2014, 15, 39. [Google Scholar] [CrossRef] [Green Version]
- Watkins, P.B.; Zimmerman, H.J.; Knapp, M.J.; I Gracon, S.; Lewis, K.W. Hepatotoxic effects of tacrine administration in patients with Alzheimer’s disease. JAMA 1994, 271, 992–998. [Google Scholar] [CrossRef]
- Gracon, S.I.; Knapp, M.J.; Berghoff, W.G.; Pierce, M.; DeJong, R.; Lobbestael, S.J.; Symons, J.; Dombey, S.L.; Luscombe, F.A.; Kraemer, D. Safety of Tacrine: Clinical Trials, Treatment IND, and Postmarketing Experience. Alzheimer Dis. Assoc. Disord. 1998, 12, 93–101. [Google Scholar] [CrossRef]
- Harrill, A.; Roach, J.; Fier, I.; Eaddy, J.S.; Kurtz, C.L.; Antoine, D.J.; Spencer, D.M.; Kishimoto, T.K.; Pisetsky, D.S.; Park, B.K.; et al. The Effects of Heparins on the Liver: Application of Mechanistic Serum Biomarkers in a Randomized Study in Healthy Volunteers. Clin. Pharmacol. Ther. 2012, 92, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.; Harrill, A.H.; Menguy-Vacheron, F.; Jayyosi, Z.; Benzerdjeb, H.; Watkins, P.B. Benign elevations in serum aminotransferases and biomarkers of hepatotoxicity in healthy volunteers treated with cholestyramine. BMC Pharmacol. Toxicol. 2014, 15, 42. [Google Scholar] [CrossRef] [Green Version]
- Rautou, P.-E.; Cazals–Hatem, D.; Moreau, R.; Francoz, C.; Feldmann, G.; Lebrec, D.; Ogier-Denis, E.; Bedossa, P.; Valla, D.; Durand, F. Acute Liver Cell Damage in Patients With Anorexia Nervosa: A Possible Role of Starvation-Induced Hepatocyte Autophagy. Gastroenterology 2008, 135, 840–848.e3. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, J.H.; Clemens, M.M.; Allard, F.D.; Yee, E.U.; Kennon-McGill, S.; Mackintosh, S.G.; Jaeschke, H.; Hambuchen, M.D.; McGill, M.R. Identification of Serum Biomarkers to Distinguish Hazardous and Benign Aminotransferase Elevations. Toxicol. Sci. 2020, 173, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Mcburney, R.N.; Hines, W.M.; Von Tungeln, L.S.; Schnackenberg, L.K.; Beger, R.D.; Moland, C.L.; Han, T.; Fuscoe, J.C.; Chang, C.-W.; Chen, J.J.; et al. The Liver Toxicity Biomarker Study: Phase I Design and Preliminary Results. Toxicol. Pathol. 2009, 37, 52–64. [Google Scholar] [CrossRef] [PubMed]
- McBurney, R.N.; Hines, W.M.; VonTungeln, L.S.; Schnackenberg, L.K.; Beger, R.D.; Moland, C.L.; Han, T.; Fuscoe, J.C.; Chang, C.-W.; Chen, J.J.; et al. The Liver Toxicity Biomarker Study Phase I: Markers for the Effects of Tolcapone or Entacapone. Toxicol. Pathol. 2012, 40, 951–964. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vazquez, J.H.; McGill, M.R. Redrawing the Map to Novel DILI Biomarkers in Circulation: Where Are We, Where Should We Go, and How Can We Get There? Livers 2021, 1, 286-293. https://doi.org/10.3390/livers1040022
Vazquez JH, McGill MR. Redrawing the Map to Novel DILI Biomarkers in Circulation: Where Are We, Where Should We Go, and How Can We Get There? Livers. 2021; 1(4):286-293. https://doi.org/10.3390/livers1040022
Chicago/Turabian StyleVazquez, Joel H., and Mitchell R. McGill. 2021. "Redrawing the Map to Novel DILI Biomarkers in Circulation: Where Are We, Where Should We Go, and How Can We Get There?" Livers 1, no. 4: 286-293. https://doi.org/10.3390/livers1040022
APA StyleVazquez, J. H., & McGill, M. R. (2021). Redrawing the Map to Novel DILI Biomarkers in Circulation: Where Are We, Where Should We Go, and How Can We Get There? Livers, 1(4), 286-293. https://doi.org/10.3390/livers1040022






