Functional and Compositional Analysis of the Fecal and Vaginal Microbiota in Vestibulodynia: An Explorative Case–Control Study
Abstract
1. Introduction
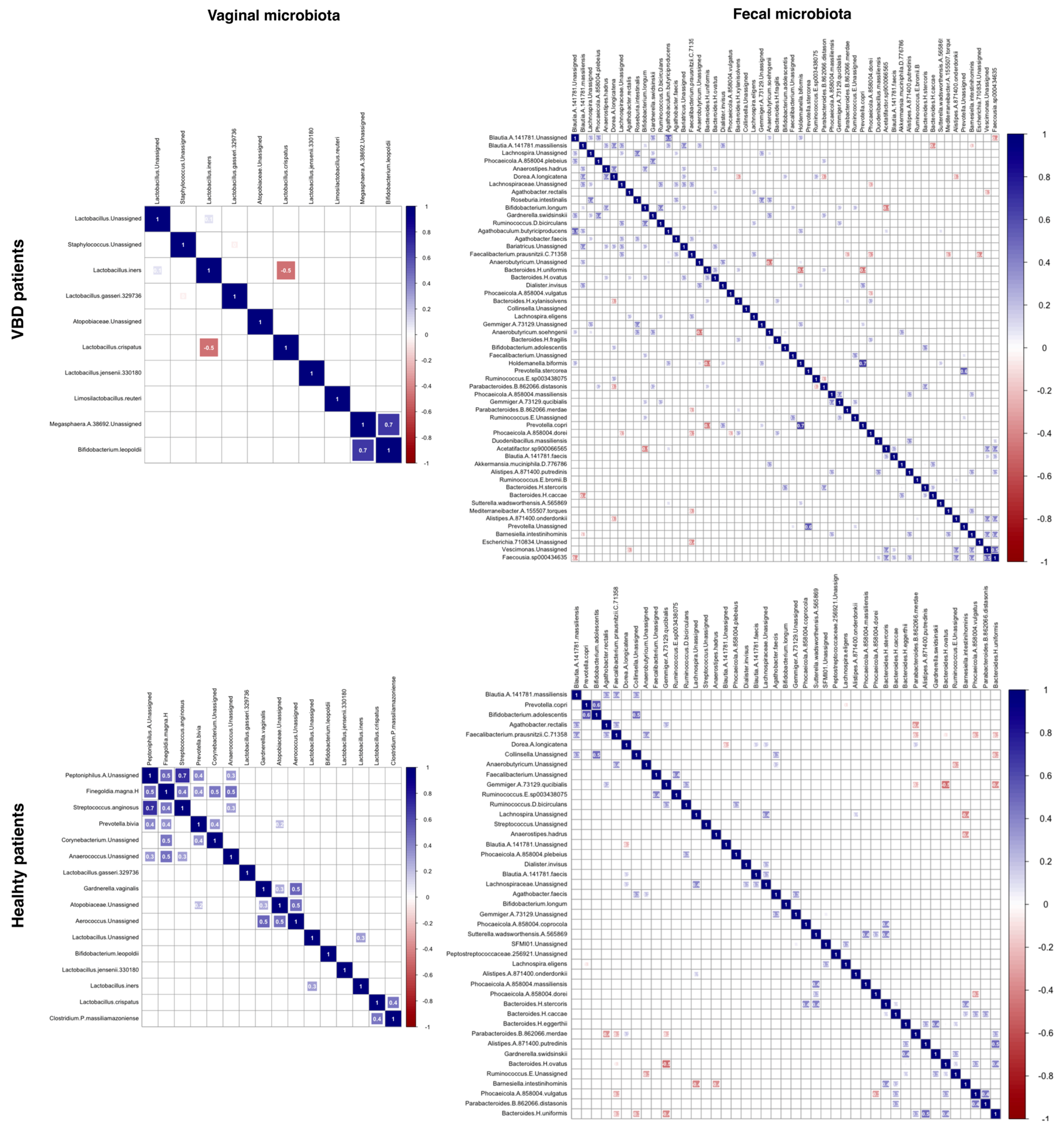
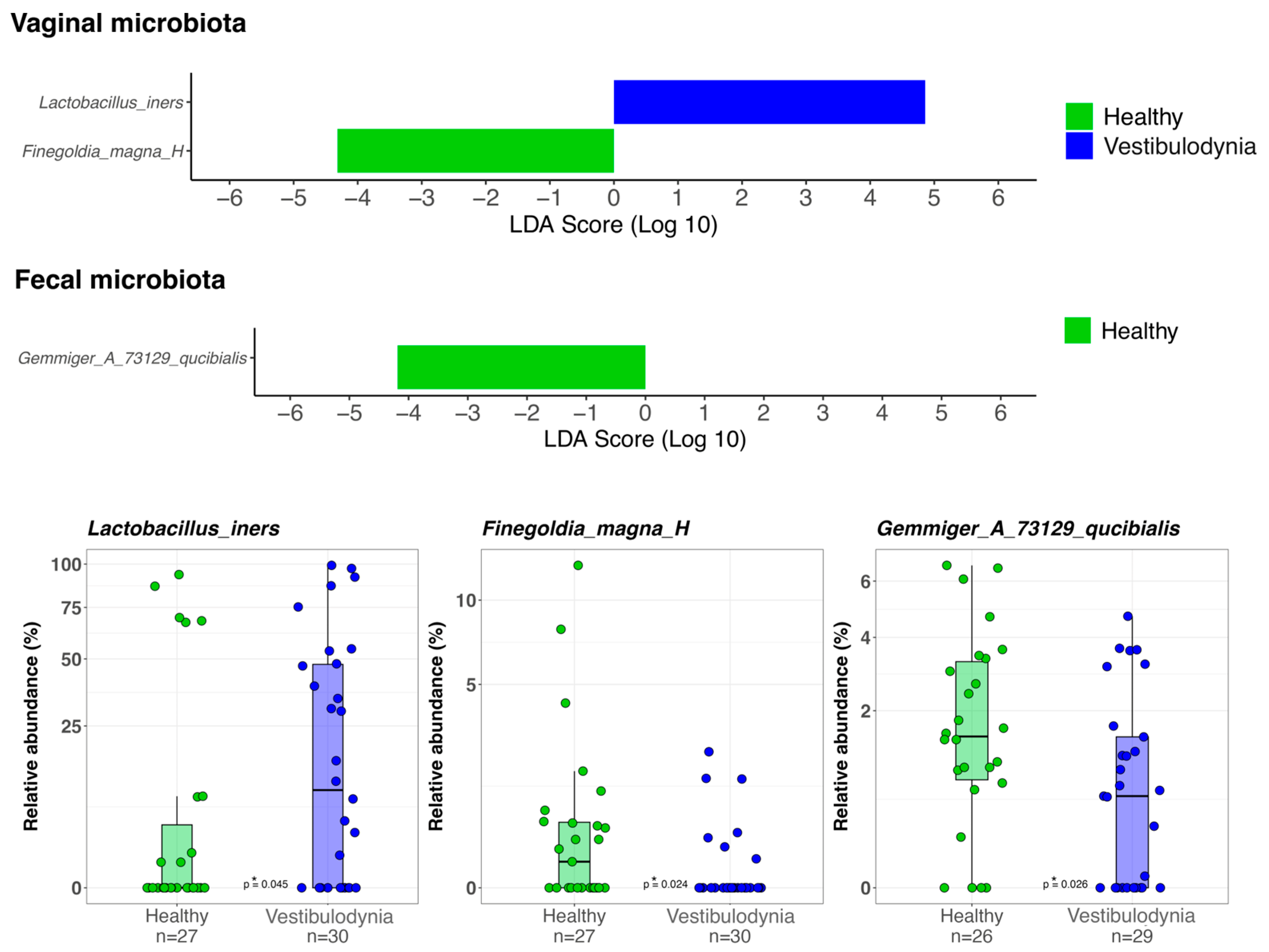
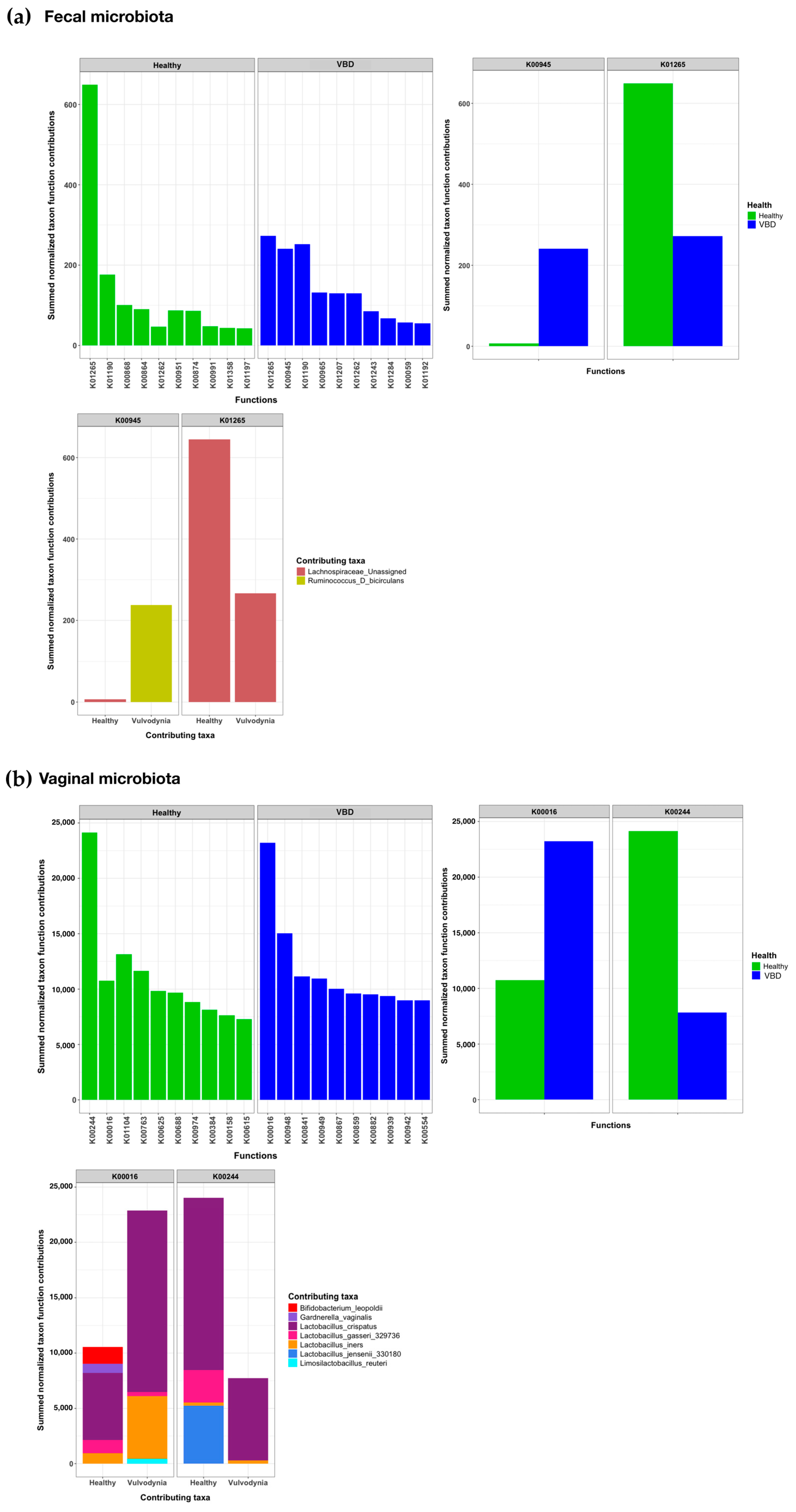
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection and Methodology
4.3. DNA Extraction and Purification
4.4. Determination of Bacterial Profiles by Amplicon Sequencing
4.5. Data Processing and Analysis
4.6. Functional Prediction of the Microbiota
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CST | Community State Type |
| VBD | Vestibulodynia |
| PCoA | Principal coordinate analysis |
| LDA LEfSe | Linear discriminant analysis effect size |
| NGS | Next-generation sequencing |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic Acid |
| L-LDH | L-lactate dehydrogenase |
| PCR | Polymerase Chain Reaction |
| FRDA | Fumarate reductase flavoprotein subunit |
Appendix A

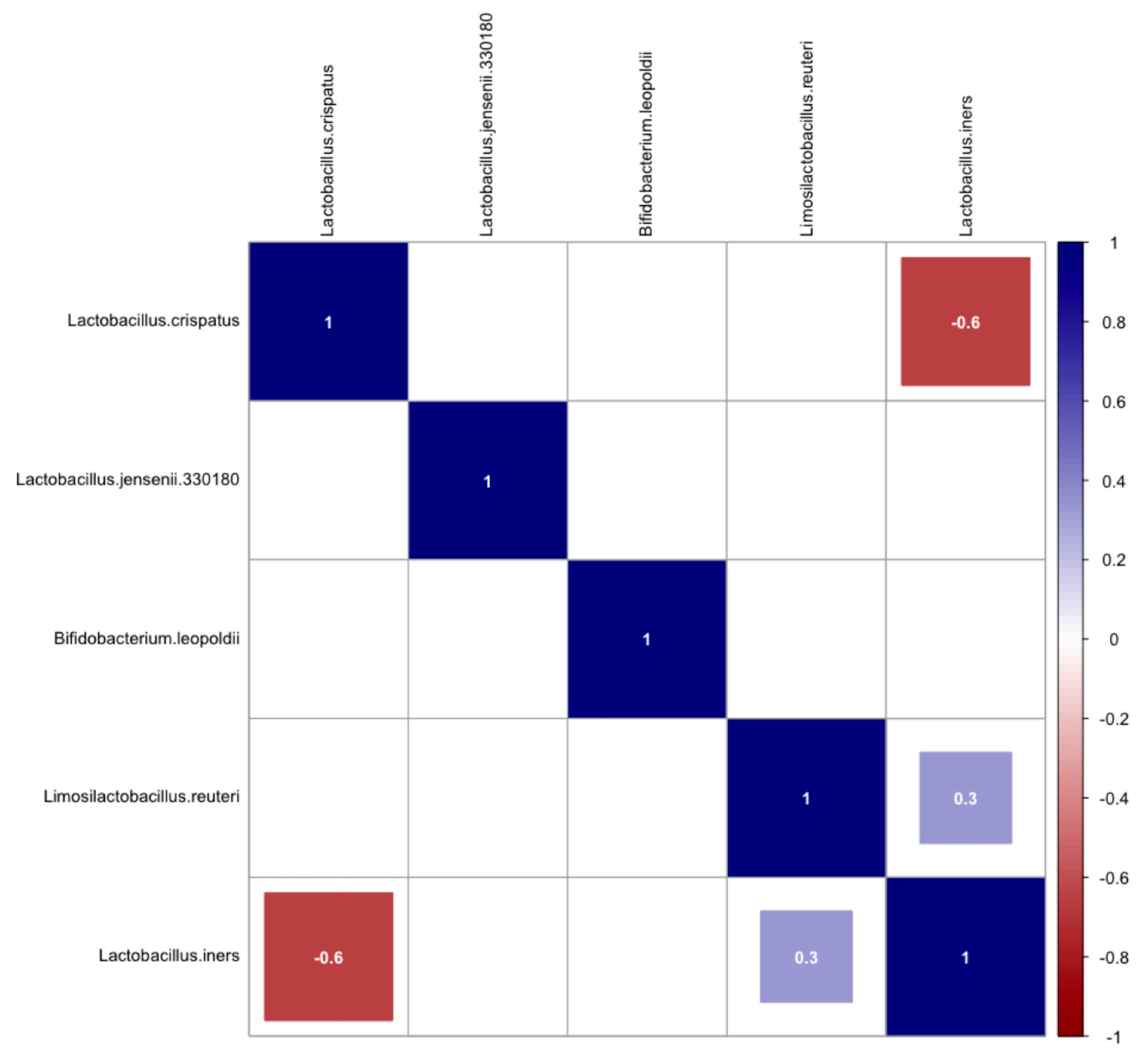

References
- De Seta, F.; Lonnee-Hoffmann, R.; Campisciano, G.; Comar, M.; Verstraelen, H.; Vieira-Baptista, P.; Ventolini, G.; Lev-Sagie, A. The Vaginal Microbiome: III. The Vaginal Microbiome in Various Urogenital Disorders. J. Low. Genit. Tract Disease 2022, 26, 85. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, J.; Goldstein, A.T.; Stockdale, C.K.; Bergeron, S.; Pukall, C.; Zolnoun, D.; Coady, D.; International Society for the Study of Vulvovaginal Disease (ISSVD). 2015 ISSVD, ISSWSH, and IPPS Consensus Terminology and Classification of Persistent Vulvar Pain and Vulvodynia. J. Sex. Med. 2016, 13, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.M.; Watson, L.; Mitchell, A.J.; Hyrien, O.; Bergerat, A.; Valint, D.J.; Pascale, A.; Hoffman, N.; Srinivasan, S.; Fredricks, D.N. Vaginal Microbiota and Mucosal Immune Markers in Women With Vulvovaginal Discomfort. Sex. Transm. Dis. 2020, 47, 269. [Google Scholar] [CrossRef] [PubMed]
- Jayaram, A.; Witkin, S.S.; Zhou, X.; Brown, C.J.; Rey, G.E.; Linhares, I.M.; Ledger, W.J.; Forney, L.J. The Bacterial Microbiome in Paired Vaginal and Vestibular Samples from Women with Vulvar Vestibulitis Syndrome. Pathog. Dis. 2014, 72, 161–166. [Google Scholar] [CrossRef]
- Bedford, L.; Parker, S.E.; Davis, E.; Salzman, E.; Hillier, S.L.; Foxman, B.; Harlow, B.L. Characteristics of the Vaginal Microbiome in Women with and without Clinically Confirmed Vulvodynia. Am. J. Obstet. Gynecol. 2020, 223, 406.e1–406.e16. [Google Scholar] [CrossRef]
- Panzarella, D.A.; Peresleni, T.; Collier, J.L.; Kocis, C.; Baker, D.A. Vestibulodynia and the Vaginal Microbiome: A Case-Control Study. J. Sex. Med. 2022, 19, 1451–1462. [Google Scholar] [CrossRef]
- Sacinti, K.G.; Razeghian, H.; Awad-Igbaria, Y.; Lima-Silva, J.; Palzur, E.; Vieira-Baptista, P.; Verstraelen, H.; Bornstein, J. Is Vulvodynia Associated With an Altered Vaginal Microbiota?: A Systematic Review. J. Low. Genit. Tract. Dis. 2024, 28, 64–72. [Google Scholar] [CrossRef]
- France, M.T.; Ma, B.; Gajer, P.; Brown, S.; Humphrys, M.S.; Holm, J.B.; Waetjen, L.E.; Brotman, R.M.; Ravel, J. VALENCIA: A Nearest Centroid Classification Method for Vaginal Microbial Communities Based on Composition. Microbiome 2020, 8, 166. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Somerfield, P.J.; Clarke, K.R.; Warwick, R.M. Simpson Index. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Oxford, UK, 2008; pp. 3252–3255. ISBN 978-0-08-045405-4. [Google Scholar]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An Effective Distance Metric for Microbial Community Comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Lev-Sagie, A.; De Seta, F.; Verstraelen, H.; Ventolini, G.; Lonnee-Hoffmann, R.; Vieira-Baptista, P. The Vaginal Microbiome: II. Vaginal Dysbiotic Conditions. J. Low. Genit. Tract Dis. 2022, 26, 79. [Google Scholar] [CrossRef]
- Tao, Z.; Zhang, L.; Zhang, Q.; Lv, T.; Chen, R.; Wang, L.; Huang, Z.; Hu, L.; Liao, Q. The Pathogenesis Of Streptococcus Anginosus In Aerobic Vaginitis. IDR 2019, 12, 3745–3754. [Google Scholar] [CrossRef] [PubMed]
- Van Hellemond, J.J.; Tielens, A.G.M. Expression and Functional Properties of Fumarate Reductase. Biochem. J. 1994, 304, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Coda, L.; Cassis, P.; Angioletti, S.; Angeloni, C.; Piloni, S.; Testa, C. Evaluation of Gut Microbiota in Patients With Vulvovestibular Syndrome. J. Clin. Med. Res. 2021, 13, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Awad-Igbaria, Y.; Palzur, E.; Nasser, M.; Vieira-Baptista, P.; Bornstein, J. Changes in the Vaginal Microbiota of Women With Secondary Localized Provoked Vulvodynia. J. Low. Genit. Tract Dis. 2022, 26, 339. [Google Scholar] [CrossRef]
- Murina, F.; Caimi, C.; Di Pierro, F.; Di Francesco, S.; Cetin, I. Features of the Vaginal and Vestibular Microbioma in Patients With Vestibulodynia: A Case-Control Study. J. Low. Genit. Tract Dis. 2020, 24, 290–294. [Google Scholar] [CrossRef]
- Galipeau, H.J.; Caminero, A.; Turpin, W.; Bermudez-Brito, M.; Santiago, A.; Libertucci, J.; Constante, M.; Raygoza Garay, J.A.; Rueda, G.; Armstrong, S.; et al. Novel Fecal Biomarkers That Precede Clinical Diagnosis of Ulcerative Colitis. Gastroenterology 2021, 160, 1532–1545. [Google Scholar] [CrossRef]
- Delgado-Diaz, D.J.; Jesaveluk, B.; Hayward, J.A.; Tyssen, D.; Alisoltani, A.; Potgieter, M.; Bell, L.; Ross, E.; Iranzadeh, A.; Allali, I.; et al. Lactic Acid from Vaginal Microbiota Enhances Cervicovaginal Epithelial Barrier Integrity by Promoting Tight Junction Protein Expression. Microbiome 2022, 10, 141. [Google Scholar] [CrossRef]
- Lauer, E.; Kandler, O. Lactobacillus Gasseri Sp. Nov., a New Species of the Subgenus Thermobacterium. Zentralblatt Für Bakteriol.: I. Abt. Orig. C Allg. Angew. Und Okol. Mikrobiol. 1980, 1, 75–78. [Google Scholar] [CrossRef]
- Witkin, S.S.; Mendes-Soares, H.; Linhares, I.M.; Jayaram, A.; Ledger, W.J.; Forney, L.J. Influence of Vaginal Bacteria and D- and L-Lactic Acid Isomers on Vaginal Extracellular Matrix Metalloproteinase Inducer: Implications for Protection against Upper Genital Tract Infections. mBio 2013, 4, e00460-13. [Google Scholar] [CrossRef] [PubMed]
- Rahkonen, L.; Rutanen, E.-M.; Unkila-Kallio, L.; Nuutila, M.; Nieminen, P.; Sorsa, T.; Paavonen, J. Factors Affecting Matrix Metalloproteinase-8 Levels in the Vaginal and Cervical Fluids in the First and Second Trimester of Pregnancy. Hum. Reprod. 2009, 24, 2693–2702. [Google Scholar] [CrossRef]
- Vieira-Baptista, P.; Bornstein, J. Candidiasis, Bacterial Vaginosis, Trichomoniasis and Other Vaginal Conditions Affecting the Vulva. In Vulvar Disease: Breaking the Myths; Bornstein, J., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 167–205. ISBN 978-3-319-61621-6. [Google Scholar]
- Rashad, A.L.; Toffler, W.L.; Wolf, N.; Thornburg, K.; Kirk, E.P.; Ellis, G.; Whitehead, W.E. Vaginal PO2 in Healthy Women and in Women Infected with Trichomonas Vaginalis: Potential Implications for Metronidazole Therapy. Am. J. Obstet. Gynecol. 1992, 166, 620–624. [Google Scholar] [CrossRef]
- O’Hanlon, D.E.; Moench, T.R.; Cone, R.A. Vaginal pH and Microbicidal Lactic Acid When Lactobacilli Dominate the Microbiota. PLoS ONE 2013, 8, e80074. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucl. Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.; Beiko, R.G. 16S rRNA Gene Analysis with QIIME2. In Microbiome Analysis: Methods and Protocols; Beiko, R.G., Hsiao, W., Parkinson, J., Eds.; Springer: New York, NY, USA, 2018; pp. 113–129. ISBN 978-1-4939-8728-3. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 Unifies Microbial Data in a Single Reference Tree. Nat. Biotechnol. 2024, 42, 715–718. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 28 April 2025).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Rbiom: Read/Write, Analyze, and Visualize “BIOM” Data. Available online: https://cmmr.r-universe.dev/rbiom (accessed on 29 April 2025).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; van den Brand, T.; et al. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics 2025. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 6 June 2025).
- Wickham, H. RStudio Tidyverse: Easily Install and Load the “Tidyverse” 2023. Available online: https://cran.r-project.org/web/packages/tidyverse/index.html (accessed on 6 June 2025).
- Wickham, H.; Vaughan, D.; Girlich, M.; Ushey, K.; Software, P. PBC Tidyr: Tidy Messy Data 2024. Available online: https://cran.r-project.org/web/packages/tidyr/index.html (accessed on 6 June 2025).
- Paradis, E.; Blomberg, S.; Bolker, B.; Brown, J.; Claramunt, S.; Claude, J.; Cuong, H.S.; Desper, R.; Didier, G.; Durand, B.; et al. Ape: Analyses of Phylogenetics and Evolution 2024. Available online: https://cran.r-project.org/web/packages/ape/index.html (accessed on 6 June 2025).
- Ggplot2 Based Publication Ready Plots. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 29 April 2025).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D.; Software, P. PBC Dplyr: A Grammar of Data Manipulation 2023. Available online: https://cran.r-project.org/web/packages/dplyr/index.html (accessed on 6 June 2025).
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple Statistical Identification and Removal of Contaminant Sequences in Marker-Gene and Metagenomics Data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package 2025. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 6 June 2025).
- Kolde, R. Pheatmap: Pretty Heatmaps 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 6 June 2025).
- R: The R Stats Package. Available online: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html (accessed on 29 April 2025).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]


| Anamnestic and Clinical Data | Control Group (N = 27) | Patients with VBD (N = 30) | p-Value |
|---|---|---|---|
| Age (average years ± SD) | 31.2 ± 5.0 | 29.4 ± 5.5 | |
| BMI (average ± SD) | 20.5 ± 1.5 | 19.8 ± 1.9 | |
| Recurring vaginitis | 0 | 13 | <0.01 |
| Contraceptive use | 21 | 20 | |
| Disease duration: 3–12 months 24–48 months 60–120 months | NA NA NA | 5 13 2 |
| CSTs | Control Group (N, %) | Patients with VBD (N, %) | p-Value |
|---|---|---|---|
| CST I | 12 (44.4) | 17 (56.7) | 0.4309 |
| CST II | 4 (14.8) | 2 (6.7) | 0.4077 |
| CST III | 5 (18.5) | 9 (30.0) | 0.3686 |
| CST IV-B | 1 (3.7) | 0 (0.0) | 0.4737 |
| CST IV-C3 | 2 (7.4) | 1 (3.3) | 0.5986 |
| CST johnsonii-dominated | 0 (0.0) | 1 (3.3) | 1.0000 |
| CST V | 3 (11.1) | 0 (0.0) | 0.1000 |
| CST I + CST III | 17 (63.0) | 26 (86.7) | 0.0631 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viciani, E.; Santacroce, B.; Padella, A.; Velichevskaya, A.; Marcante, A.; Di Rito, L.; Soverini, M.; Graziottin, A.; Murina, F.; Castagnetti, A. Functional and Compositional Analysis of the Fecal and Vaginal Microbiota in Vestibulodynia: An Explorative Case–Control Study. Women 2025, 5, 22. https://doi.org/10.3390/women5030022
Viciani E, Santacroce B, Padella A, Velichevskaya A, Marcante A, Di Rito L, Soverini M, Graziottin A, Murina F, Castagnetti A. Functional and Compositional Analysis of the Fecal and Vaginal Microbiota in Vestibulodynia: An Explorative Case–Control Study. Women. 2025; 5(3):22. https://doi.org/10.3390/women5030022
Chicago/Turabian StyleViciani, Elisa, Barbara Santacroce, Antonella Padella, Alena Velichevskaya, Andrea Marcante, Laura Di Rito, Matteo Soverini, Alessandra Graziottin, Filippo Murina, and Andrea Castagnetti. 2025. "Functional and Compositional Analysis of the Fecal and Vaginal Microbiota in Vestibulodynia: An Explorative Case–Control Study" Women 5, no. 3: 22. https://doi.org/10.3390/women5030022
APA StyleViciani, E., Santacroce, B., Padella, A., Velichevskaya, A., Marcante, A., Di Rito, L., Soverini, M., Graziottin, A., Murina, F., & Castagnetti, A. (2025). Functional and Compositional Analysis of the Fecal and Vaginal Microbiota in Vestibulodynia: An Explorative Case–Control Study. Women, 5(3), 22. https://doi.org/10.3390/women5030022






