Ruminal Lipid A Analysis by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Ruminal LPS Extraction
2.3. Ruminal Lipid A Isolation
2.4. MALDI-TOF MS Analysis
2.5. DNA Extraction, PCR Amplification, and 16S rRNA Sequencing
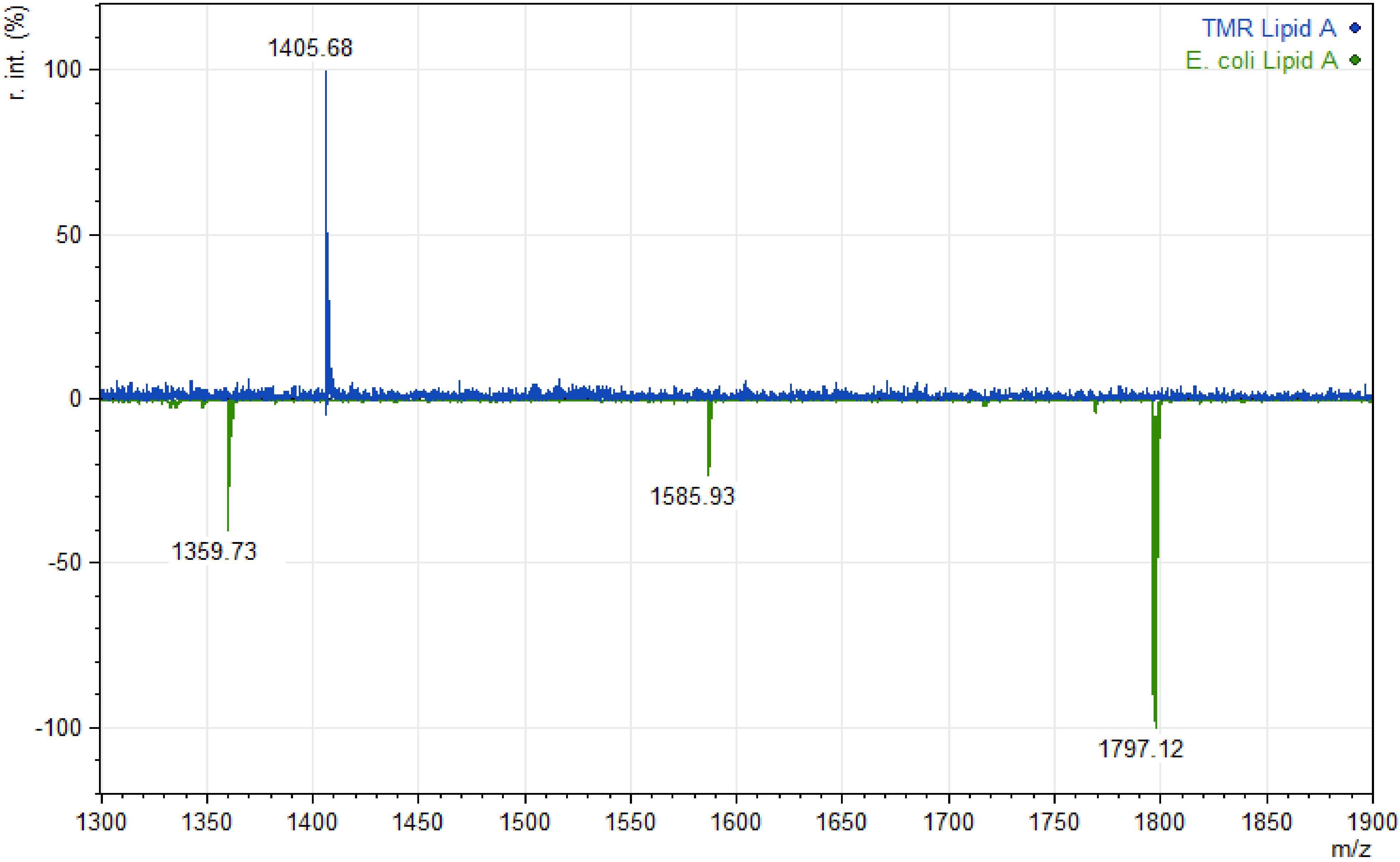
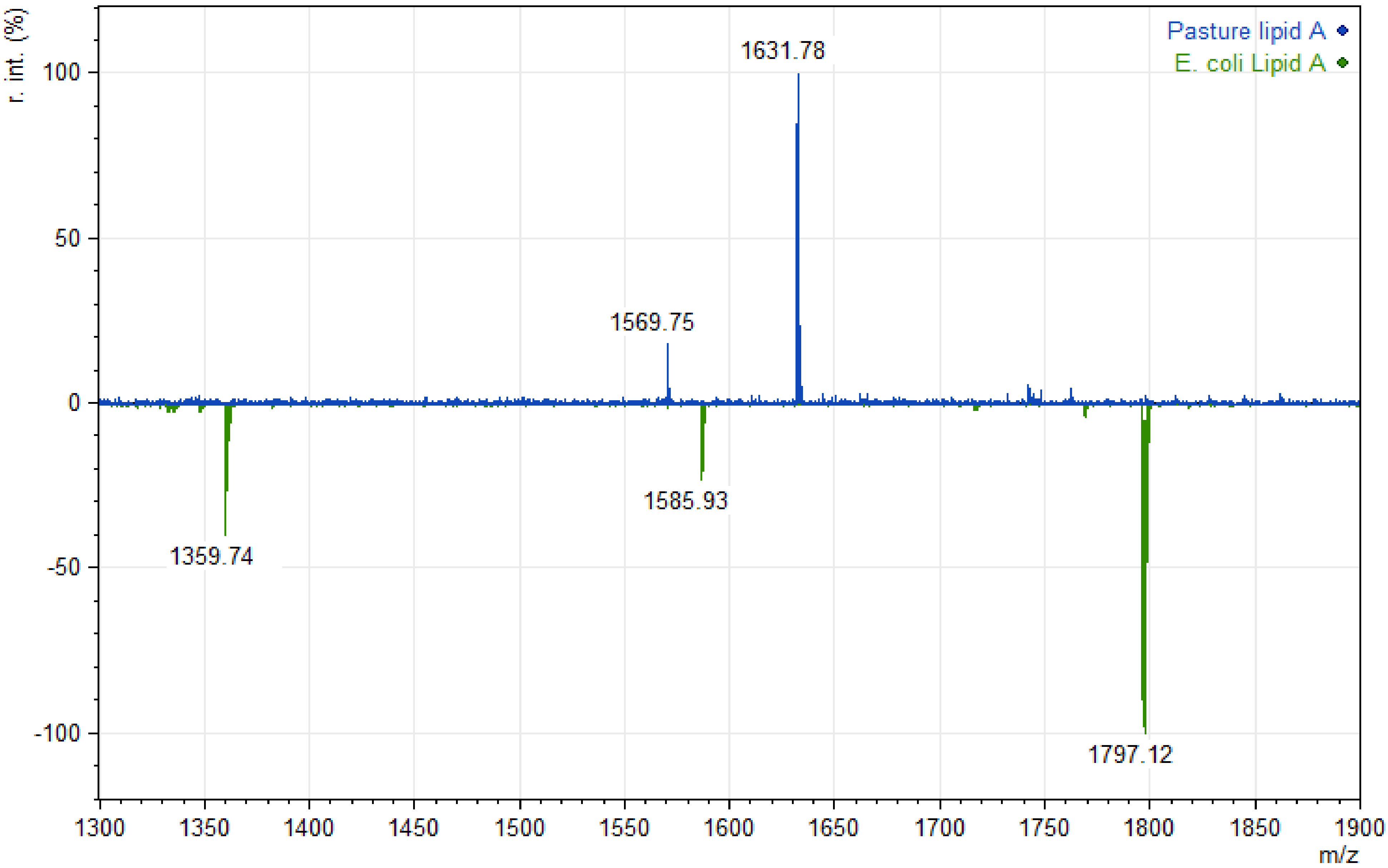
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [Google Scholar] [CrossRef]
- Erridge, C.; Bennett-Guerrero, E.; Poxton, I.R. Structure and function of lipopolysaccharides. Microbes Infect. 2002, 4, 837–851. [Google Scholar] [CrossRef]
- Lodowska, J.; Wolny, D.; Weglarz, L.; Dzierzewicz, Z. The structural diversity of lipid A from gram-negative bacteria. Postep. Hig. Med. Dosw. 2007, 61, 106–121. [Google Scholar]
- Coats, S.R.; Pham, T.-T.T.; Bainbridge, B.W.; Reife, R.A.; Darveau, R.P. MD-2 Mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 2005, 175, 4490–4498. [Google Scholar] [CrossRef] [Green Version]
- Munford, R.S.; Varley, A.W. Shield as signal: Lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2006, 2, e67. [Google Scholar] [CrossRef]
- Steimle, A.; Autenrieth, I.B.; Frick, J.-S. Structure and function: Lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol. 2016, 306, 290–301. [Google Scholar] [CrossRef] [Green Version]
- Nagaraja, T.G.; Lechtenberg, K.F. Acidosis in feedlot cattle. Vet. Clin. N. Am. Food Anim. Pract. 2007, 23, 333–350. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Li, S.; Danscher, A.M.; Derakshani, H.; Andersen, P.H.; Khafipour, E. Changes in microbiota in rumen digesta and feces due to a grain-based subacute ruminal acidosis (SARA) challenge. Microb. Ecol. 2017, 74, 485–495. [Google Scholar] [CrossRef]
- Khafipour, E.; Li, S.; Plaizier, J.C.; Krause, D.O. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microbiol. 2009, 75, 7115–7124. [Google Scholar] [CrossRef] [Green Version]
- Nagaraja, T.G.; Bartley, E.E.; Fina, L.R.; Anthony, H.D.; Bechtle, R.M. Evidence of endotoxins in the rumen bacteria of cattle fed hay or grain. J. Anim. Sci. 1978, 47, 226–234. [Google Scholar] [CrossRef]
- D’Hennezel, E.; Abubucker, S.; Murphy, L.O.; Cullen, T.W. Total lipopolysaccharide from the human gut microbiome silences toll-like receptor signaling. mSystems 2017, 2, e00046-17. [Google Scholar] [CrossRef] [Green Version]
- Sonesson, H.R.A.; Zähringer, U.; Grimmecke, H.D.; Westphal, O.R.E. Bacterial endotox. In Chemical Structure, Biological, Activity; Bringham, K., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1994. [Google Scholar]
- Berezow, A.B.; Ernst, R.K.; Coats, S.R.; Braham, P.H.; Karimi-Naser, L.M.; Darveau, R.P. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb. Pathog. 2009, 47, 68–77. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, A.N.; Choudhury, B.P.; Fischbach, M.A. The biosynthesis of lipooligosaccharide from Bacteroides thetaiotaomicron. mBio 2018, 9, e02289-17. [Google Scholar] [CrossRef] [Green Version]
- Bilal, M.S.; Abaker, J.A.; Ul Aabdin, Z.; Xu, T.; Dai, H.; Zhang, K.; Liu, X.; Shen, X. Lipopolysaccharide derived from the digestive tract triggers an inflammatory response in the uterus of mid-lactating dairy cows during SARA. BMC Vet. Res. 2016, 12, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizsan, S.J.; Ahvenjärvi, S.; Volden, H.; Broderick, G.A. Estimation of rumen outflow in dairy cows fed grass silage-based diets by use of reticular sampling as an alternative to sampling from the omasal canal1. J. Dairy Sci. 2010, 93, 1138–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, J.S.; Anderson, E.M.; Hao, Y. LPS quantitation procedures. Methods Mol. Biol. 2014, 1149, 375–402. [Google Scholar] [CrossRef]
- Zhou, P.; Altman, E.; Perry, M.B.; Li, J. Study of matrix additives for sensitive analysis of lipid A by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 2010, 76, 3437–3443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Paula, E.M.; Lelis, A.L.J.; Silva, L.G.; Brandao, V.L.N.; Monteiro, H.F.; Fan, P.; Poulson, S.R.; Jeong, K.C.; Faciola, A.P. Effects of lipopolysaccharide dosing on bacterial community composition and fermentation in a dual-flow continuous culture system. J. Dairy Sci. 2019, 102, 334–350. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.W.; Shaffer, S.A.; Ernst, R.K.; Goodlett, D.R.; Turecek, F. Determination of pyrophosphorylated forms of lipid A in gram-negative bacteria using a multivaried mass spectrometric approach. Proc. Nat. Acad. Sci. USA 2008, 105, 12742–12747. [Google Scholar] [CrossRef] [Green Version]
- De Menezes, A.B.; Lewis, E.; O’Donovan, M.; O’Neill, B.F.; Clipson, N.; Doyle, E.M. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 2011, 78, 256–265. [Google Scholar] [CrossRef] [Green Version]
- Sarmikasoglou, E.; Faciola, A.P. Ruminal lipopolysaccharides analysis: Uncharted waters with promising signs. Animals 2021, 11, 195. [Google Scholar] [CrossRef]
- Li, Y.; Powell, D.A.; Shaffer, S.A.; Rasko, D.A.; Pelletier, M.R.; Leszyk, J.D.; Scott, A.J.; Masoudi, A.; Goodlett, D.R.; Wang, X.; et al. LPS remodeling is an evolved survival strategy for bacteria. Proc. Nat. Acad. Sci. USA 2012, 109, 8716–8721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuffey, R.K.; Richardson, L.F.; Wilkinson, J.I.D. Ionophores for dairy cattle: Current status and future outlook. J. Dairy Sci. 2001, 84, E194–E203. [Google Scholar] [CrossRef]
| Phyla in Ruminal Content | Proportion of Sequences | |
|---|---|---|
| Total Mixed Ration 1 | Pasture | |
| Bacteroidetes | 63.32 | 28.46 |
| Firmicutes | 23.82 | 34.29 |
| Spirochaetes | 5.79 | 12.26 |
| Proteobacteria | 2.75 | 23.78 |
| Fibrobacteres | 1.84 | 0.00 |
| Euryarchaeota | 1.10 | 0.16 |
| Synergistetes | 0.65 | 0.00 |
| Planctomycetes | 0.43 | 0.19 |
| Cyanobacteria | 0.30 | 0.50 |
| Kiritimatiellaeota | not detected | 0.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmikasoglou, E.; Vinyard, J.R.; Khan, M.S.; Jiranantasak, T.; Ravelo, A.; Lobo, R.R.; Fan, P.; Jeong, K.C.; Tuanyok, A.; Faciola, A. Ruminal Lipid A Analysis by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Polysaccharides 2021, 2, 817-824. https://doi.org/10.3390/polysaccharides2040049
Sarmikasoglou E, Vinyard JR, Khan MS, Jiranantasak T, Ravelo A, Lobo RR, Fan P, Jeong KC, Tuanyok A, Faciola A. Ruminal Lipid A Analysis by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Polysaccharides. 2021; 2(4):817-824. https://doi.org/10.3390/polysaccharides2040049
Chicago/Turabian StyleSarmikasoglou, Efstathios, James R. Vinyard, Mohamed S. Khan, Treenate Jiranantasak, Anay Ravelo, Richard R. Lobo, Peixin Fan, Kwangcheol C. Jeong, Apichai Tuanyok, and Antonio Faciola. 2021. "Ruminal Lipid A Analysis by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry" Polysaccharides 2, no. 4: 817-824. https://doi.org/10.3390/polysaccharides2040049
APA StyleSarmikasoglou, E., Vinyard, J. R., Khan, M. S., Jiranantasak, T., Ravelo, A., Lobo, R. R., Fan, P., Jeong, K. C., Tuanyok, A., & Faciola, A. (2021). Ruminal Lipid A Analysis by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Polysaccharides, 2(4), 817-824. https://doi.org/10.3390/polysaccharides2040049







