GC-MS-Based Metabolomics Analysis of Prawn Shell Waste Co-Fermentation by Lactobacillus plantarum and Bacillus subtilis
Abstract
1. Introduction
2. Materials and Methods
2.1. Fermentation Conditions and Harvesting of Samples
2.2. Samples Preparation for Extracellular Metabolites Analysis
2.3. GC-MS Analysis of Extracellular Metabolites
2.4. Statistical Analysis of Metabolites
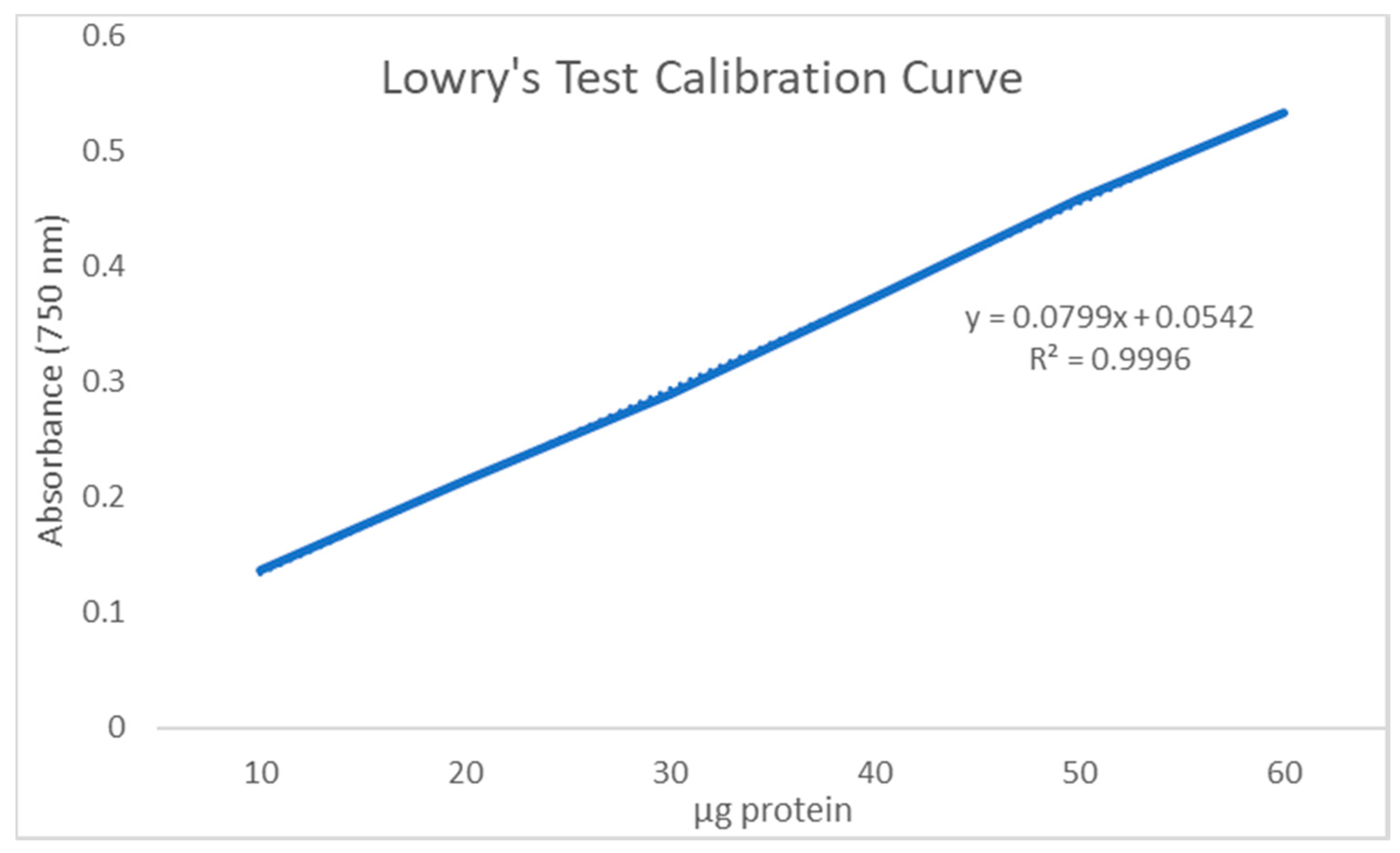
2.5. Determination of Chitin Yield and Purity
3. Results
3.1. Metabolomics Analysis by GC-MS
3.2. Chitin Yield and Purity Calculations
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kandra, P.; Challa, M.M.; Jyothi, H.K.P. Efficient use of shrimp waste: Present and future trends. Appl. Microbiol. Biotechnol. 2012, 93, 17–29. [Google Scholar] [CrossRef]
- Ferrer, J.; Paez, G.; Marmol, Z.; Ramones, E.; Garcia, H.; Forster, C.F. Acid hydrolysis of shrimp-shell wastes and the production of single cell protein from the hydrolysate. Bioresour. Technol. 1996, 57, 55–60. [Google Scholar] [CrossRef]
- Gomez-Rios, D.; Barrera-Zapata, R.; Rios-Estepa, R. Comparison of process technologies for chitosan production from shrimp shell waste: A techno-economic approach using Aspen Plus. Food Bioprod. Process. 2017, 103, 49–57. [Google Scholar] [CrossRef]
- Manni, L.; Ghorbel-Bellaaj, O.; Jellouli, K.; Younes, I.; Nasri, M. Extraction and Characterization of Chitin, Chitosan, and Protein Hydrolysates Prepared from Shrimp Waste by Treatment with Crude Protease from Bacillus cereus SV1. Appl. Biochem. Biotechnol. 2010, 162, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Arancibia, M.Y.; Aleman, A.; Calvo, M.M.; Lopez-Caballero, M.E.; Montero, P.; Gomez-Guillen, M.C. Antimicrobial and antioxidant chitosan solutions enriched with active shrimp (Litopenaeus vannamei) waste materials. Food Hydrocoll. 2014, 35, 710–717. [Google Scholar] [CrossRef]
- Mao, X.; Liu, P.; He, S.; Xie, J.; Kan, F.; Yu, C.; Li, Z.; Xue, C.; Lin, H. Antioxidant Properties of Bio-active Substances from Shrimp Head Fermented by Bacillus licheniformis OPL-007. Appl. Biochem. Biotechnol. 2013, 171, 1240–1252. [Google Scholar] [CrossRef] [PubMed]
- Benhabiles, M.S.; Abdi, N.; Drouiche, N.; Lounici, H.; Pauss, A.; Goosen, M.F.A.; Mameri, N. Protein recovery by ultrafiltration during isolation of chitin from shrimp shells Parapenaeus longirostris. Food Hydrocoll. 2013, 32, 28–34. [Google Scholar] [CrossRef]
- El-Beltagy, A.E.; El-Sayed, S.M. Functional and nutritional characteristics of protein recovered during isolation of chitin from shrimp waste. Food Bioprod. Process. 2012, 90, 633–638. [Google Scholar] [CrossRef]
- Prameela, K.; Venkatesh, K.; Immandi, S.B.; Katsuri, A.P.K.; Krishna, C.R.; Mohan, C.M. Next generation nutraceutical from shrimp waste: The convergence of applications with extraction methods. Food Chem. 2017, 237, 121–132. [Google Scholar] [CrossRef]
- Robert, M.; Zatylyn-Gaudin, C.; Fournier, V.; Corre, E.; Corguille, G.L.; Bernay, B.; Henry, J. Transcriptomic and peptidomic analysis of protein hydrolysates from the white shrimp (L. vannamei). J. Biotechnol. 2014, 186, 30–37. [Google Scholar] [CrossRef]
- Bueno-Solano, C.; Lopez-Cervantes, J.; Campas-Baypoli, O.N.; Lauterio-Garcia, R.; Adan-Bante, N.P.; Sanchez-Machado, D.I. Chemical and biological characteristics of protein hydrolysates from fermented shrimp by-products. Food Chem. 2009, 112, 671–675. [Google Scholar] [CrossRef]
- Sila, A.; Nasri, M.; Bougatef, A. Isolation and characterization of carotenoproteins from deep-water pink shrimp processing waste. Int. J. Biol. Macromol. 2012, 51, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Narayan, B.; Velappan, S.P.; Zituji, S.P.; Manjabhatta, S.N.; Gowda, L.R. Yield and chemical composition of fractions from fermented shrimp biowaste. Waste Manag. Res. 2010, 28, 64–70. [Google Scholar] [CrossRef]
- Perez-Santin, E.; Calvo, M.M.; Lopez-Caballero, M.E.; Montero, P.; Gomez-Guillen, M.C. Compositional properties and bioactive potential of waste material from shrimp cooking juice. LWT Food Sci. Technol. 2013, 54, 87–94. [Google Scholar] [CrossRef]
- Sachindra, N.M.; Bhaskar, N. In vitro antioxidant activity of liquor from fermented shrimp biowaste. Bioresour. Technol. 2008, 99, 9013–9016. [Google Scholar] [CrossRef]
- Armenta-Lopez, R.; Guerrero, L.I.; Huerta, S. Astaxanthin Extraction from Shrimp Waste by Lactic Acid Fermentation and Enzymatic Hydrolysis of the Carotenoprotein Complex. Food Chem. Toxicol. 2002, 67, 1002–1006. [Google Scholar]
- Gomez-Estaca, J.; Calvo, M.M.; Alvarez-Acero, I.; Montero, P.; Gomez-Guillen, M.C. Characterization and storage stability of astaxanthin esters, fatty acid profile and α-tocopherol of lipid extract from shrimp (L. vannamei) waste with potential applications as food ingredient. Food Chem. 2017, 216, 37–44. [Google Scholar] [CrossRef]
- Sila, A.; Sayari, N.; Balti, R.; Martinez-Alvarez, O.; Nedjar-Arroume, N.; Moncef, N.; Bougatef, A. Biochemical and antioxidant properties of peptidic fraction of carotenoproteins generated from shrimp by-products by enzymatic hydrolysis. Food Chem. 2014, 148, 445–452. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, J.; Kan, F.; Gao, Y.; Lan, J.; Zhang, X.; Hu, Z.; Li, Y.; Lin, H. Antioxidant Production and Chitin Recovery from Shrimp Head Fermentation with Streptococcus thermophilus. Food Sci. Biotechnol. 2013, 22, 1023–1032. [Google Scholar] [CrossRef]
- Francisco, F.C.; Simora, R.M.C.; Nunal, S.N. Deproteination and demineralization of shrimp waste using lactic acid bacteria for the production of crude chitin and chitosan. Aquac. Bioflux 2015, 8, 107–115. [Google Scholar]
- Maruthiah, T.; Somanath, B.; Immanuel, G.; Palavesam, A. Deproteinization potential and antioxidant property of haloalkalophilic organic solvent protease from marine Bacillus sp. APCMST-RS3 using marine shell wastes. Biotechnol. Rep. 2015, 8, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Armenta, R.E.; Guerrero-Legarreta, I. Amino acid profile and enhancement of the enzymatic hydrolysis of fermented shrimp carotenoproteins. Food Chem. 2009, 112, 310–315. [Google Scholar] [CrossRef]
- Jung, W.J.; Jo, G.H.; Kuk, J.H.; Kim, K.Y.; Park, R.D. Extraction of chitin from red crab shell waste by cofermentation with Lactobacillus paracasei subsp. tolerans KCTC-3074 and Serratia marcescens FS-3. Appl. Microbiol. Biotechnol. 2006, 71, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Aytekin, O.; Elibol, M. Cocultivation of Lactococcus lactis and Teredinobacter turnirae for biological chitin extraction from prawn waste. Bioprocess. Biosyst. Eng. 2010, 33, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Phuong, P.T.D.; Minh, N.C.; Cuong, H.N.; Minh, N.V.; Han, N.T.; Hoa, N.V.; Yen, H.T.H.; Trung, T.S. Recovery of protein hydrolysate and chitosan from black tiger shrimp (Penaeus monodon) heads: Approaching a zero waste process. J. Food Sci. Technol. 2017, 54, 1850–1856. [Google Scholar] [CrossRef] [PubMed]
- Fernando, R.; Munasinghe, D.M.S.; Gunasena, A.R.C.; Abeynayake, P. Determination of nitrofuran metabolites in shrimp muscle by liquid chromatography-photo diode array detection. Food Control 2017, 72, 300–305. [Google Scholar] [CrossRef]
- Chen, D.; Ye, Y.; Chen, J.; Yan, X. Evolution of metabolomics profile of crab paste during fermentation. Food Chem. 2016, 192, 886–892. [Google Scholar] [CrossRef]
- Xiao, M.; Qian, K.; Wang, Y.; Bao, F. GC-MS metabolomics reveals metabolic differences of the farmed Mandarin fish Siniperca chuatsi in recirculating ponds aquaculture system and pond. Sci. Rep. 2020, 10, 6090. [Google Scholar] [CrossRef]
- Ma, Q.Q.; Wang, X.D.; Cui, Y.Y.; Zhang, N.N.; Qin, J.G.; Du, Z.Y.; Chen, L.Q. Untargeted GC-MS metabolomics reveals metabolic differences in the Chinese mitten-hand crab (Eriocheir sinensis) fed with dietary palm oil or olive oil. Aquacult. Nutr. 2018, 24, 1623–1637. [Google Scholar] [CrossRef]
- Zang, J.; Xu, Y.; Xia, W.; Jiang, Q.; Yang, F.; Wang, B. Phospholipid molecular species composition of Chinese traditional low-salt fermented fish inoculated with different starter cultures. Food Res. Int. 2018, 111, 87–96. [Google Scholar] [CrossRef]
- Ming, T.; Han, J.; Li, Y.; Lu, C.; Qiu, D.; Li, Y.; Zhou, J.; Su, X. A metabolomics and proteomics study of the Lactobacillus plantarum in the grass carp fermentation. BMC Microbiol. 2018, 18, 216. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Cervantes, J.; Sanchez-Machado, D.I.; Rosas-Rodriguez, J.A. Analysis of free amino acids in fermented shrimp waste by high-performance liquid chromatography. J. Chromatogr. A 2005, 1105, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Cervantes, J.; Sanchez-Machado, D.I.; Rosas-Rodriguez, J.A. High-performance liquid chromatography method for the simultaneous quantification of retinol, α-tocopherol, and cholesterol in shrimp waste hydrolysate. J. Chromatogr. A 2006, 1105, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Park, S.E.; Yoo, S.A.; Seo, S.H.; Lee, K.I.; Na, C.S.; Son, H.S. GC-MS based metabolomics approach of Kimchi for the understanding of Lactobacillus plantarum fermentation characteristics. LWT Food Sci. Technol. 2016, 68, 313–321. [Google Scholar] [CrossRef]
- Yang, X.; Hu, W.; Xiu, Z.; Jiang, A.; Yang, X.; Sarengaowa; Ji, Y.; Guan, Y.; Feng, K. Microbial dynamics and volatilome profiles during the fermentation of Chinese northeast sauerkraut by Leuconostoc mesenteroides ORC 2 and Lactobacillus plantarum HBUAS 51041 under different salt concentrations. Food Res. Int. 2020, 130, 108926. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kang, J.; Ma, Z.; Li, X.; Liu, L.; Hu, X. Microbial succession and metabolite changes during traditional serofluid dish fermentation. LWT Food Sci. Technol. 2017, 84, 771–779. [Google Scholar] [CrossRef]
- Li, P.; Tang, H.; Shi, C.; Xie, Y.; Zhou, H.; Xia, B.; Zhang, C.; Chen, L.; Jiang, L. Untargeted metabolomics analysis of Mucor racemosus Douchi fermentation by gas chromatography with time-of-flight mass spectrometry. Food Sci. Nutr. 2019, 7, 1865–1874. [Google Scholar] [CrossRef]
- Srivastava, S.; Kumar, P.R.; Mishra, S.K. Identification of Metabolites through GC/LC-MS Processed Data using Different Reference Libraries and Their Comparison. J. Pharm. Biomed. Sci. 2016, 6, 363–368. [Google Scholar]
- Devi, R.; Dhamodharan, R. Pretreatment in Hot Glycerol for Facile and Green Separation of Chitin from Prawn Shell Waste. ACS Sustain. Chem. Eng. 2018, 6, 846–853. [Google Scholar] [CrossRef]
- Toche, R.B.; Janrao, R.A. Synthesis and characterization and antimicrobial evaluation of novel urea, sulfonamide and acetamide 3,4-dihydropyrazinol[1,2-a]indol-1(2H)-one derivates. Arab. J. Chem. 2019, 12, 3406–3416. [Google Scholar] [CrossRef]
- Popiolek, L.; Biernasiuk, A. Design, synthesis, and in vitro antimicrobial activity of hydrazide-hydrazones of 2-substituted acetic acid. Chem. Bio. Drug Des. 2016, 88, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Fraise, A.P.; Wilkinson, M.A.C.; Bradley, C.R.; Oppenheim, B.; Moiemen, N. The antibacterial activity and stability of acetic acid. J. Hosp. Infect. 2013, 84, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Josephrajan, T.; Ramakrishnan, V.T.; Kathiravan, G.; Muthumary, J. Synthesis and antimicrobial studies of some acridinediones and their thiourea derivatives. Arkivoc 2005, 11, 124–136. [Google Scholar]
- Gratzl, G.; Paulik, C.; Hild, S.; Guggenbichler, J.P.; Lackner, M. Antimicrobial activity of poly(acrylic acid) block copolymers. Mater. Sci. Eng. C 2014, 38, 94–100. [Google Scholar] [CrossRef]
- Meeta, M.; Kumar, P.; Narasimhan, B. Synthesis, antimicrobial evaluation and QSAR studies of p-amino benzoic acid derivatives. J. Pharm. Technol. Res. Manag. 2014, 2, 339–356. [Google Scholar]
- Sariguney, A.B.; Kocabas, E.; Erci, F.; Torlak, E.; Coskun, A. Synthesis and Antimicrobial Activity of Some 2-aminothiazole and 2-aminothiadiazine Derivatives. J. Heterocycl. Chem. 2018, 55, 2107–2110. [Google Scholar] [CrossRef]
- Das, U.N. Arachidonic acid and other unsaturated fatty acids and some of their metabolites function as endogenous antimicrobial molecules: A review. J. Adv. Res. 2018, 11, 57–66. [Google Scholar] [CrossRef]
- Leong, H.J.; Oh, S.G. Preparation of antibacterial TiO2 particles by hybridization with azelaic acid for applications in cosmetics. J. Ind. Eng. Chem. 2018, 66, 242–247. [Google Scholar] [CrossRef]
- Nuta, D.C.; Chifiriuc, M.C.; Draghici, C.; Limban, C.; Missir, A.V.; Morusciag, L. Synthesis, characterization and antimicrobial activity evaluation of new agents from benzamides class. Farmacia 2013, 61, 966–974. [Google Scholar]
- Kim, M.G.; Lee, H.S. 1,2-benzenediol isolated from persimmon roots and its structural analogues show antimicrobial activities against food-borne bacteria. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 429–433. [Google Scholar] [CrossRef]
- Abdelkader, M.S.A.; Rateb, M.E.; Mohamed, G.A.; Jaspars, M. Harpulliasides A and B: Two new benzeneacetic acid derivatives from Harpullia pendula. Phytochem. Lett. 2016, 15, 131–135. [Google Scholar] [CrossRef]
- Li, J.; Duan, M.; Yao, X.; Tian, D.; Tang, J. Prenylated benzenepropanoic acid analogues from the Citrus grandis (L.) Osbeck and their anti-neuroinflammatory activity. Fitoterapia 2019, 139, 104410. [Google Scholar] [CrossRef] [PubMed]
- Igwe, C.N.; Okoro, U.C. Synthesis, Characterization and Evaluation for Antibacterial and Antifungal Activites of N-Heteroaryl Substituted Benzene Sulphonamides. Org. Chem. Int. 2014. [Google Scholar] [CrossRef]
- Ambala, A.; Lincoln, C.A. Synthesis, characterization, antimicrobial activity and DNA cleavage study of (E)-2-(((2-(P-Tolyloxy)Quinolin-3-Yl)Methylene)Amino)Benzenethiol Schiff base metal complexes. Chem. Data Collect. 2020, 27. [Google Scholar] [CrossRef]
- Vicini, P.; Zani, F.; Cozzini, P.; Doytchinova, I. Hydrazones of 1,2-benzisothiazole hydrazides: Synthesis, antimicrobial activity and QSAR investigations. Eur. J. Med. Chem. 2002, 37, 553–564. [Google Scholar] [CrossRef]
- Alex, D.; Gay-Andrieu, F.; May, J.; Thampi, L.; Dou, D.; Mooney, A.; Groutas, W.; Calderone, R. Amino Acid-Derived 1,2-Benzisothiazolinone Derivatives as Novel Small-Molecule Antifungal Inhibitors: Identification of Potential Genetic Targets. Antimicrob. Agents Chemother. 2012, 56, 4630–4639. [Google Scholar] [CrossRef]
- Rakesh, K.P.; Shantharam, C.S.; Sridhara, M.B.; Manukumar, H.M.; Qin, H.L. Benzisoxazole: A privileged scaffold for medicinal chemistry. Med. Chem. Commun. 2017, 8, 2023–2039. [Google Scholar] [CrossRef]
- Chen, H.; Zhong, Q. Antibacterial activity of acidified sodium benzoate against Escherichia coli O157:H7, Salmonella enterica, and Listeria monocytogenes in tryptic soy broth and on cherry tomatoes. Int. J. Food Microbiol. 2018, 274, 38–44. [Google Scholar] [CrossRef]
- Leite, A.C.L.; da Silva, K.P.; de Souza, I.A.; de Araujo, J.M.; Brondani, D.J. Synthesis, antitumour and antimicrobial activities of new peptidyl derivatives containing the 1,3-benzodioxole system. Eur. J. Med. Chem. 2004, 39, 1059–1065. [Google Scholar] [CrossRef]
- Park, E.S.; Moon, W.S.; Song, M.J.; Kim, M.N.; Chung, K.H.; Yoon, J.S. Antimicrobial activity of phenol and benzoic acid derivatives. Int. Biodeterior. Biodegrad. 2001, 47, 209–214. [Google Scholar] [CrossRef]
- Carcamo-Noriega, E.N.; Sathyamoorthi, S.; Banerjee, S.; Gnanamani, E.; Mendoza-Trujillo, M.; Mata-Espinosa, D.; Hernandez-Pando, R.; Veytia-Bucheli, J.I.; Possani, L.D.; Zare, R.N. 1,4-Benzoquinone antimicrobial agents against Staphylococcus aureus and Mycobacterium tuberculosis derived from scorpion venom. Proc. Natl. Acad. Sci. USA 2019, 116, 12642–12647. [Google Scholar] [CrossRef] [PubMed]
- Rida, S.M.; Ashour, F.A.; El-Hawash, S.A.M.; El-Semary, M.M.; Badr, M.H.; Shalaby, M.A. Synthesis of some novel benzoxazole derivatives as anticancer, anti-HIV-1 and antimicrobial agents. Eur. J. Med. Chem. 2005, 40, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Kashid, A.M.; Dube, P.N.; Alkutkar, P.G.; Bothara, K.G.; Mokale, S.N.; Dhawale, S.C. Synthesis, biological screening and ADME prediction of benzylindole derivatives as novel anti-HIV-1, anti-fungal and anti-bacterial agents. Med. Chem. Res. 2013, 22, 4633–4640. [Google Scholar] [CrossRef]
- Gouda, K.G.M.; Kavitha, M.D.; Sarada, R. Antihyperglycemic, antioxidant and antimicrobial activities of the butanol extract from Spirulina Platensis. J. Food Biochem. 2015, 39, 594–602. [Google Scholar] [CrossRef]
- Namkung, H.; Yu, H.; Gong, J.; Leeson, S. Antimicrobial activity of butyrate glycerides towards Salmonella Typhimurium and Clostridium perfringens. Poultr. Sci. 2011, 90, 2217–2222. [Google Scholar] [CrossRef]
- Kratky, M.; Vinsova, J. Salicylanilide N-monosubstituted carbamates: Synthesis and in vitro antimicrobial activity. Bioorg. Med. Chem. 2016, 24, 1322–1330. [Google Scholar] [CrossRef]
- Zanatta, N.; Borchhardt, D.M.; Alves, S.H.; Coelho, H.S.; Squizani, A.M.C.; Marchi, T.M.; Bonacorso, H.G.; Martins, M.A.P. Synthesis and antimicrobial activity of new (4,4,4-trihalo-3-oxo-but-1-enyl)-carbamic acid ethyl esters, (4,4,4-trihalo-3-hydroxy-butyl)-carbamic acid ethyl esters, and 2-oxo-6-trihalomethyl-[1,3]oxazinane-3-carboxylic acid ethyl esters. Bioorg. Med. Chem. 2006, 14, 3174–3184. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Jain, S.; Kumar, G. Synthesis, antimicrobial evaluation, QSAR and in silico ADMET studies of decanoic acid derivatives. Pol. Pharm. Soc. Drug Res. 2011, 68, 191–204. [Google Scholar]
- Al-Dhabi, N.A.; Arasu, M.V.; Rejiniemon, T.S. In Vitro Antibacterial, Antifungal, Antibiofilm, Antioxidant, and Anticancer Properties of Isosteviol Isolated from Endangered Medicinal Plant Pittosporum tetraspermum. Evid.-Based Complem. Altern. Med. 2015, 2015, 164261. [Google Scholar] [CrossRef]
- Shin, S.Y.; Bajpai, V.K.; Kim, H.R.; Kang, S.C. Antibacterial activity of bioconverted eicosapentaenoic (EPA) and docosahexaenoic acid (DHA) against foodborne pathogenic bacteria. Int. J. Food Microbiol. 2007, 113, 233–236. [Google Scholar] [CrossRef]
- Lakshmi, S.A.; Bhaskar, J.P.; Krishnan, V.; Sethupathy, S.; Pandipriya, S.; Aruni, W.; Pandian, S.K. Inhibition of biofilm and biofilm-associated virulence factor production in methicillin-resistant Staphylococcus aureus by docosanol. J. Biotechnol. 2020, 317, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Abdalha, A.A.; Mekawey, A.A.I. Antimicrobial Susceptibility of Certain Fungal and Bacterial Strains to Dodecanamide and Quinazolinone Derivatives. World Appl. Sci. J. 2013, 24, 312–319. [Google Scholar]
- Oh, D.H.; Marshall, D.L. Antimicrobial activity of ethanol, glycerol monolaurate or lactic acid against List. Monocytogenes. Int. J. Food Microbiol. 1993, 20, 239–246. [Google Scholar] [CrossRef]
- Tamokou, J.D.D.; Mpetga, D.J.S.; Lunga, P.K.; Tene, M.; Tane, P.; Kuiate, J.R. Antioxidant and antimicrobial activities of ethyl acetate extract, fractions and compounds from stem bark of Albizia adianthifolia (Mimosoideae). Bmc Complem. Altern. Med. 2012, 12. [Google Scholar] [CrossRef]
- Huisjes, E.H.; de Hulster, E.; van Dam, J.C.; Pronk, J.T.; van Maris, A.J.A. Galacturonic Acid Inhibits the Growth of Saccharomyces cerevisiae on Galactose, Xylose, and Arabinose. Appl. Environ. Microbiol. 2012, 78, 5052–5059. [Google Scholar] [CrossRef]
- Nieto-Penalver, C.G.; Savino, M.J.; Bertini, E.V.; Sanchez, L.A.; de Figueroa, L.I.C. Gluconic acid produced by Gluconacetobacter diazotrophicus Pal5 possesses antimicrobial properties. Res. Microbiol. 2014, 165, 549–558. [Google Scholar] [CrossRef]
- Berger, F.M.; Hubbard, C.V.; Ludwig, B.J. The Antimicrobial Action of Certain Glycerol Ethers and Related Compounds. Appl. Microbiol. 1953, 1, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Nurullaeva, M.K.; Azizov, U.M.; Mikhlina, E.E.; Turchin, K.F.; Silin, V.A.; Yakhontov, L.N. Synthesis of (3-pyridyl)glyoxylic acid derivatives and their antimicrobial properties. Pharmaceut. Chem. J. 1987, 20, 563–567. [Google Scholar] [CrossRef]
- Liu, H.; Lepoittevin, B.; Roddier, C.; Guerineau, V.; Bech, L.; Herry, J.M.; Bellon-Fontaine, M.N.; Roger, P. Facile synthesis and promising antibacterial properties of a new guaiacol-based polymer. Polymer 2011, 52, 1908–1916. [Google Scholar] [CrossRef]
- Vasudevan, A.; Vijayan, D.; Mandal, P.; Haridas, M. Anti-inflammatory Property of n-Hexadecanoic Acid: Structural Evidence and Kinetic Assessment. Chem. Biol. Drug Des. 2010, 9, 1236–1240. [Google Scholar]
- Narasimhan, B.; Judge, V.; Narang, R.; Ohlan, R.; Ohlan, S. Quantitative structure-activity relationship studies for prediction of antimicrobial activity of synthesized 2,4-hexadienoic acid derivatives. Bioorg. Med. Chem. Lett. 2007, 17, 5835–5845. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.H.; Jiang, M.H. Evaluation of antibacterial activity of hexanedioic acid isolated from Hermetia illucens larvae. J. Appl. Biomed. 2014, 12, 179–189. [Google Scholar] [CrossRef]
- Alva-Murillo, N.; Ochoa-Zarzosa, A.; Lopez-Meza, J.E. Short chain fatty acids (propionic and hexanoic) decrease Staphylococcus aureus internalization into bovine mammary epithelial cells and modulate antimicrobial peptide expression. Vet. Microbiol. 2012, 155, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Kratky, M.; Vinsova, J.; Volkova, M.; Buchta, V.; Trejtnar, F.; Stolarikova, J. Antimicrobial activity of sulfonamides containing 5-chloro-2-hydroxybenzaldehyde and 5-chloro-2-hydroxybenzoic acid scaffold. Eur. J. Med. Chem. 2012, 50, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, Z.; Li, J.; Yang, X.; Fei, B.; Leung, P.H.M.; Tao, X. A New Antimicrobial Agent: Poly (3-hydroxybutyric acid) Oligomer. Macromol. Biosci. 2019, 19. [Google Scholar] [CrossRef]
- Kumar, A.; Mishra, A.K. Synthesis and antimicrobial activity of some new diphenylamine derivatives. J. Pharm. Bioallied Sci. 2015, 7, 81–85. [Google Scholar] [CrossRef]
- Sakko, M.; Moore, C.; Novak-Fraser, L.; Rautemaa, V.; Sorsa, T.; Hietala, P.; Jarvinen, A.; Bowyer, P.; Tjaderhane, L.; Rautemaa, R. 2-hydroxyisocaproic acid is fungicidal for Candida and Aspergillus species. Mycoses 2014, 57, 214–221. [Google Scholar] [CrossRef]
- Sundar, L.; Chang, F.N. Antimicrobial activity and biosynthesis of indole antibiotics produced by Xenorhabdus nematophilus. J. Gen. Microbiol. 1993, 139, 3139–3148. [Google Scholar] [CrossRef]
- Himaja, M.; Jose, T.; Ramana, M.V.; Anand, R.; Munirajasekhar, D. Synthesis and biological evaluation of indole-3-carboxylic acid derivatives of amino acids and peptides. Int. Res. J. Pharm. 2010, 1, 436–440. [Google Scholar]
- In, Y.W.; Kim, J.J.; Kim, H.J.; Oh, S.W. Antimicrobial activities of acetic acid, citric acid and lactic acid against Shigella species. J. Food Saf. 2013, 33, 79–85. [Google Scholar] [CrossRef]
- Ferrazzano, L.; Viola, A.; Lonati, E.; Bulbarelli, A.; Musumeci, R.; Cocuzza, C.; Lombardo, M.; Tolomelli, A. New isoxazolidinone and 3,4-dehydro-β-proline derivatives as antibacterial agents as MAO-inhibitors: A complex balance between two activities. Eur. J. Med. Chem. 2010, 124, 906–919. [Google Scholar] [CrossRef] [PubMed]
- Velikova, M.; Bankova, V.; Tsvetkova, I.; Kujumgiev, A.; Marcucci, M.C. Antibacterial ent-kaurene from Brazilian propolis of native stingless bees. Fitoterapia 2000, 71, 693–696. [Google Scholar] [CrossRef]
- Wang, C.; Chang, T.; Yang, H.; Cui, M. Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella Enteritidis, Escherichia coli and Listeria monocytogenes. Food Control 2015, 47, 231–236. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, Y.S.; Shin, D.H. Antimicrobial Synergistic Effect of Linolenic Acid and Monoglyceride against Bacillus cereus and Staphylococcus aureus. J. Agric. Food Chem. 2002, 50, 2193–2199. [Google Scholar] [CrossRef]
- Orhan, I.; Ozeelik, B.; Aslan, S.; Kartal, M.; Karaoglu, T.; Sener, B.; Terzioglu, S.; Choudhary, M.I. Antioxidant and antimicrobial actions of the clubmoss Lycopodium clavatum L. Phytochem. Rev. 2007, 6, 189–196. [Google Scholar] [CrossRef]
- Raybaudi-Massilia, R.M.; Mosqueda-Melgar, J.; Martin-Belloso, O. Antimicrobial activity of malic acid against Listeria monocytogenes, Salmonella Enteritidis and Escherichia coli O157:H7 in apple, pear and melon juices. Food Control 2009, 20, 105–112. [Google Scholar] [CrossRef]
- Motamedifar, M.; Bazargani, A.; Namazi, M.R.; Sarai, H.S.E. Antimicrobial Activity of Mandelic Acid Against Methicillin-Resistant Staphylococcus aureus: A Novel Finding with Important Practical Implications. World Appl. Sci. J. 2014, 31, 925–929. [Google Scholar]
- Ristovski, J.T.; Jankovic, N.; Borcic, V.; Jain, S.; Bugarcic, Z.; Mikov, M. Evaluation of antimicrobial activity and retention behavior of newly synthesized vanilidene derivatives of Meldrum’s acids using QSRR approach. J. Pharm. Biomed. Anal. 2018, 155, 42–49. [Google Scholar] [CrossRef]
- Karaman, I.; Sahin, F.; Gulluce, M.; Ogutcu, H.; Sengul, M.; Adiguzel, A. Antimicrobial activity of aqueous and methanol extracts of Juniperus oxycedrus L. J. Ethnopharmacol. 2003, 85, 231–235. [Google Scholar] [CrossRef]
- Kern, E.R.; Kushner, N.L.; Hartline, C.B.; Williams-Aziz, S.L.; Harden, E.A.; Zhou, S.; Zemlicka, J.; Prichard, M.N. In Vitro Activity and Mechanism of Action of Methylenecyclopropane Analogs of Nucleosides against Herpesvirus Replication. Antimicrob. Agents Chemother. 2005, 49, 1039–1045. [Google Scholar] [CrossRef]
- Yoo, J.C.; Han, J.M.; Nam, S.K.; Ko, O.H.; Choi, C.H.; Kee, K.H.; Sohng, J.K.; Jo, J.S.; Seong, C.N. Characterization and Cytotoxic Activities of Nonadecanoic Acid Produced by Streptomyces scabiei subsp. chosunensi M0137 (KCTC 9927). J. Microbiol. 2002, 40, 331–334. [Google Scholar]
- Sahin, N.; Kula, I.; Erdogan, Y. Investigation of Antimicrobial Activities of Nonanoic Acid Derivatives. Fresenius Environ. Bull. 2006, 15, 141–143. [Google Scholar]
- Pu, Z.H.; Zhang, Y.Q.; Yin, Z.Q.; Xu, J.; Jia, R.Y.; Lu, Y.; Yang, F. Antibacterial Activity of 9-Octadecanoic Acid-Hexadecanoic Acid-Tetrahydrofuran-3,4-Diyl Ester from Neem Oil. Agric. Sci. China 2010, 9, 1236–1240. [Google Scholar] [CrossRef]
- Hilgren, J.D.; Salverda, J.A. Antimicrobial Efficacy of a Peroxyacetic/Octanoic Acid Mixture in Fresh-Cut-Vegetable Process Waters. J. Food Sci. 2000, 65, 1376–1379. [Google Scholar] [CrossRef]
- De Lucena, J.M.V.M.; Decker, E.M.; Walter, C.; Boeira, L.S.; Lost, C.; Weiger, R. Antimicrobial effectiveness of intracanal medicaments on Enterococcus faecalis: Chlorhexidine versus octenidine. Int. Endod. J. 2013, 46, 53–61. [Google Scholar] [CrossRef]
- Kuhrt, M.F.; Fancher, M.J.; McKinlay, M.A.; Lennert, S.D. Virucidal Activity of Glutaric Acid and Evidence for Dual Mechanism of Action. Antimicrob. Agents Chemother. 1984, 26, 924–927. [Google Scholar] [CrossRef]
- Roy, S.; Hagen, K.D.; Maheswari, P.U.; Lutz, M.; Spek, A.L.; Reedijk, J.; van Wezel, G.P. Phenanthroline Derivatives with Improved Selectivity as DNA-Targeting Anticancer or Antimicrobial Drugs. ChemMedChem 2008, 3, 1427–1434. [Google Scholar] [CrossRef]
- Mazimba, O.; Wale, K.; Loeto, D.; Kwape, T. Antioxidant and antimicrobial studies on fused-ring pyrazolones and isoxazolones. Bioorg. Med. Chem. 2014, 22, 6564–6569. [Google Scholar] [CrossRef]
- Arias-Moliz, M.T.; Ferrer-Luque, C.M.; Espigares-Rodriguez, E.; Liebana-Urena, J.; Espigares-Garcia, M. Bactericidal activity of phosphoric acid, citric acid, and EDTA solutions against Enterococcus faecalis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2008, 106, 84–89. [Google Scholar] [CrossRef]
- Evren, A.E.; Yurttas, L.; Yilmaz-Cankilic, M. Synthesis of novel N-(naphthalen-1-yl)propanamide derivatives and evaluation of their antimicrobial activity. Phosphorussulfur Silicon Relat. Elem. 2020, 195, 158–164. [Google Scholar] [CrossRef]
- Desai, N.C.; Pandya, D.; Vaja, D. Synthesis, characterization and antimicrobial studies on 3-((4-(4-nitrophenyl)-6-aryl-1,6-dihydropyrimindin-2-yl)thio)propanenitriles and their derivatives. Med. Chem. Res. 2017, 26, 1089–1097. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, W.; Jiang, X.; Jiang, F.; Zhuang, H.; Fu, L. Design, synthesis and antimicrobial activity of chiral 2-(substituted-hydroxyl)-3-(benzo[d]oxazol-5-yl)propanoic acid derivates. Eur. J. Med. Chem. 2011, 46, 3639–3650. [Google Scholar] [CrossRef] [PubMed]
- Helal, M.H.; Abbas, S.Y.; Salem, M.H.; Farag, A.A.; Ammar, Y.A. Synthesis and characterization of new types of 2-(6-methoxy-2-naphthyl)propionamide derivatives as potential antibacterial and antifungal agents. Med. Chem. Res. 2013, 22, 5598–5609. [Google Scholar] [CrossRef]
- El Azab, I.H.; Khalifa, M.E.; Gobouri, A.A.; Altalhi, T.A. Synthesis, Characterization and Pharmacological Evaluation of Some New Pteridine-Based Heterocycles as Antimicrobial Agents. J. Heterocycl. Chem. 2019, 56, 1352–1361. [Google Scholar] [CrossRef]
- Elaasser, M.M.; Abdel-Aziz, M.M.; El-Kassas, R.A. Antioxidant, antimicrobial, antiviral and antitumour activities of pyranone derivative obtained from Aspergillus candidus. J. Microbiol. Biotech. Res. 2011, 1, 5–17. [Google Scholar]
- Premkumar, T.; Govindarajan, S. Antimicrobial study of pyrazine, pyrazole and imidazole carboxylic acids and their hydrazinium salts. World J. Microbiol. Biotechnol. 2005, 21, 479–480. [Google Scholar] [CrossRef]
- Kandile, N.G.; Mohamed, M.I.; Zaky, H.; Mohamed, H.M. Novel pyridazine derivatives: Synthesis and antimicrobial activity evaluation. Eur. J. Med. Chem. 2009, 44, 1989–1996. [Google Scholar] [CrossRef]
- Yurttas, L.; Ozkay, Y.; Kaplancikli, Z.A.; Tunali, Y.; Karaca, H. Synthesis and antimicrobial activity of some new hydrazone-bridged thiazole-pyrrole derivatives. J. Enzym. Inhib. Med. Chem. 2013, 28, 830–835. [Google Scholar] [CrossRef]
- Jacobson, J.G.; Renau, T.E.; Nassiri, M.R.; Sweier, D.G.; Breitenbach, J.M.; Townsend, L.B.; Drach, J.C. Nonnucleoside Pyrrolopyrimidines with a Unique Mechanism of Action against Human Cytomegalovirus. Antimicrob. Agents Chemother. 1999, 43, 1888–1894. [Google Scholar] [CrossRef]
- Purohit, A.; Mohan, A. Antimicrobial effects of pyruvic and succinic acids on Salmonella survival in ground chicken. LWT Food Sci. Technol. 2019, 116, 108596. [Google Scholar] [CrossRef]
- Jafari, E.; Khajouei, M.R.; Hassanzadeh, F.; Hakimelahi, G.H.; Khodarahmi, G.A. Quinazolinone and quinazoline derivatives: Recent structures with potent antimicrobial and cytotoxic activities. Res. Pharm. Sci. 2016, 11, 1–14. [Google Scholar] [PubMed]
- Eswaran, S.; Adhikari, A.V.; Shetty, N.S. Synthesis and antimicrobial activities of novel quinoline derivatives carrying 1,2,4-triazole moiety. Eur. J. Med. Chem. 2009, 44, 4637–4647. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.R. Quinolone Molecular Structure-Activity Relationships: What We Have Learned about improving Antimicrobial Activity. Clin. Infect. Dis. 2001, 33, S180–S186. [Google Scholar] [CrossRef]
- Brilisauer, K.; Rapp, J.; Rath, P.; Schollhorn, A.; Bleul, L.; Weiβ, E.; Stahl, M.; Grond, S.; Forchhammer, K. Cyanobacterial antimetabolite 7-deoxy-sedoheptulose blocks the shikimate pathway to inhibit the growth of prototrophic organisms. Nat. Commun. 2019, 10, 545. [Google Scholar] [CrossRef] [PubMed]
- Hans, S.; Sharma, S.; Hameed, S.; Fatima, Z. Sesamol exhibits potent antimycobacterial activity: Underlying mechanisms and impact on virulence traits. J. Glob. Antimicrob. Resist. 2017, 10, 228–237. [Google Scholar] [CrossRef]
- Kenawy, E.R.; Abdel-Hay, F.I.; Shahada, L.; El-Shanshoury, A.E.R.; El-Newehy, M.H. Biologically Active Polymers. IV. Synthesis and Antimicrobial Activity of Tartaric Acid Polyamides. J. Appl. Polym. Sci. 2006, 102, 4780–4790. [Google Scholar] [CrossRef]
- Bondock, S.; Fadaly, W.; Metwally, M.A. Synthesis and antimicrobial activity of some new thiazole, thiophene and pyrazole derivatives containing benzothiazole moiety. Eur. J. Med. Chem. 2010, 45, 3692–3701. [Google Scholar] [CrossRef]
- Saeed, S.; Rashid, N.; Jones, P.G.; Ali, M.; Hussain, R. Synthesis, characterization and biological evaluation of some thiourea derivatives bearing benzothiazole moiety as potential antimicrobial and anticancer agents. Eur. J. Med. Chem. 2010, 45, 1323–1331. [Google Scholar] [CrossRef]
- Guarda, A.; Rubilar, J.F.; Miltz, J.; Galotto, M.J. The antimicrobial activity of microencapsulated thymol and carvacrol. Int. J. Food Microbiol. 2011, 146, 144–150. [Google Scholar] [CrossRef]
- Bayrak, H.; Demirbas, A.; Karaoglu, S.A.; Demirbas, N. Synthesis of some new 1,2,4-triazoles, their Mannich and Schiff bases and evaluation of their antimicrobial activities. Eur. J. Med. Chem. 2009, 44, 1057–1066. [Google Scholar] [CrossRef]
- Dolezalova, M.; Janis, R.; Svobodova, H.; Kasparkova, V.; Humpolicek, P.; Krejci, J. Antimicrobial properties of 1-monoacylglycerols prepared from undecanoic (C11:0) and undecanoic (C11:1) acid. Eur. J. Lipid Sci. Technol. 2010, 112, 1106–1114. [Google Scholar] [CrossRef]
- Grether-Beck, S.; Felsner, I.; Brenden, H.; Kohne, Z.; Majora, M.; Marini, A.; Jaenicke, T.; Rodriguez-Martin, M.; Trullas, C.; Hupe, M.; et al. Urea Uptake Enhances Barrier Function and Antimicrobial Defense in Humans by Regulating Epidermal Gene Expression. J. Investig. Dermatol. 2012, 132, 1561–1572. [Google Scholar] [CrossRef] [PubMed]
- Kawsar, S.M.A.; Islam, M.; Jesmin, S.; Manchur, M.A.; Hasan, I.; Rajia, S. Evaluation of the antimicrobial activity and cytotoxic effect of some uridine derivatives. Int. J. Biosci. 2018, 12, 211–219. [Google Scholar]
- Wheatley, R.E. The consequences of volatile organic compound mediated bacterial and fungal interactions. Antonie Van Leeuwenhoek 2002, 81, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Audrain, B.; Farag, M.A.; Ryu, C.M.; Ghigo, J.M. Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol. Rev. 2015, 39, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahilon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Reis, J.A.; Paula, A.T.; Casarotti, S.N.; Penna, A.L.B. Lactic acid bacteria Antimicrobial Compounds: Characteristics and Applications. Food Eng. Rev. 2012, 4, 124–140. [Google Scholar] [CrossRef]
- Niku-Paavola, M.L.; Laitila, A.; Mattila-Sandholm, T.; Haikara, A. New types of antimicrobial compounds produced by Lactobacillus plantarum. J. Appl. Microbiol. 1999, 86, 29–35. [Google Scholar] [CrossRef]
- Siedler, S.; Balti, R.; Neves, A.R. Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr. Opin. Biotechnol. 2019, 56, 138–146. [Google Scholar] [CrossRef]
| Metabolite | Molecular Formula | Quantity (mg/L) | Biological Characteristic |
|---|---|---|---|
| Alanine | C3H7NO2 | 4642.67 ± 3.90 | Amino acid |
| Alanylthreonine | C7H14N2O4 | 26.39 ± 0.01 | Amino acid |
| Alloisoleucine | C6H13NO2 | 13.86 ± 0.01 | Amino acid |
| 1,4-Dihydrophenylalanine | C9H13NO2 | 2.92 ± 0.01 | Amino acid |
| Glutamic acid | C5H9NO4 | 5.68 ± 0.01 | Amino acid |
| Glycine | C2H5NO2 | 25.56 ± 0.02 | Amino acid |
| Isoleucine | C6H13NO2 | 13.96 ± 0.06 | Amino acid |
| Ketoisocaproic acid | C6H10O3 | 44.06 ± 0.13 | Leucine ketoacid |
| Ketoisovaleric acid | C5H8O3 | 1.20 ± 0.01 | Valine ketoacid |
| Leucine | C6H13NO2 | 63.91 ± 0.01 | Amino acid |
| Lysine | C6H14N2O2 | 0.44 ± 0.01 | Amino acid |
| Proline | C5H9NO2 | 91.76 ± 1.28 | Amino acid |
| Threonine | C4H9NO3 | 91.73 ± 0.05 | Amino acid |
| Valine | C5H11NO2 | 3.13 ± 0.01 | Amino acid |
| Metabolite | Molecular Formula | Quantity (mg/L) | Biological Characteristic |
|---|---|---|---|
| Acetamide | C2H5NO | 2999.12 ± 4.06 | Antimicrobial [40] |
| Acethydrazide | C2H6N2O | 366.71 ± 0.01 | Antimicrobial [41] |
| Acetic acid | C2H4O2 | 71.94 ± 0.17 | Antimicrobial [42] |
| Acridinedione | C13H7NO2 | 4.31 ± 0.01 | Antimicrobial [43] |
| Acrylic acid | C3H4O2 | 0.72 ± 0.01 | Antimicrobial [44] |
| Allonic acid | C6H12O7 | 18.89 ± 0.08 | Anti-tumor |
| 4-Aminobenzoic acid | C7H7NO2 | 7.00 ± 0.01 | Antimicrobial [45] |
| 2-Aminothiadiazole | C3H4N2S | 20.98 ± 0.01 | Antimicrobial [46] |
| Arachidonic acid | C20H32O2 | 5.92 ± 0.01 | Antimicrobial [47] |
| Azelaic acid | C9H16O4 | 0.50 ± 0.01 | Antimicrobial [48] |
| Benzamide | C7H7NO | 11.18 ± 0.12 | Antimicrobial [49] |
| 1,2-Benzenediol | C6H6O2 | 1.31 ± 0.01 | Antimicrobial [50] |
| Benzeneacetic acid | C8H8O2 | 9.04 ± 0.01 | Antimicrobial [51] |
| Benzenepropanoic acid | C9H10O2 | 0.84 ± 0.01 | Antimicrobial [52] |
| Benzenesulfonamide | C6H7NO2S | 167.36 ± 1.56 | Antimicrobial [53] |
| Benzenethiol | C6H6S | 1.47 ± 0.01 | Antimicrobial [54] |
| 1,2-Benzisothiazole | C7H5NS | 56.13 ± 0.59 | Antimicrobial [55] |
| Benzisothiazolinone | C7H5NOS | 23.64 ± 0.01 | Antimicrobial [56] |
| 1,2-Benzisoxazole | C7H5NO | 1.75 ± 0.01 | Antimicrobial [57] |
| Benzoate | C7H5O2− | 24.82 ± 0.01 | Antimicrobial [58] |
| 1,3-Benzodioxole | C7H6O2 | 6.97 ± 0.01 | Antimicrobial [59] |
| Benzoic acid | C7H6O2 | 27.37 ± 0.38 | Antimicrobial [60] |
| 1,4-Benzoquinone | C6H4O2 | 28.12 ± 0.37 | Antimicrobial [61] |
| Benzoxazole | C7H5NO | 1.26 ± 0.01 | Antimicrobial [62] |
| 1-Benzylindole | C15H13N | 7.92 ± 0.04 | Antimicrobial [63] |
| Butanol | C4H10O | 1.81 ± 0.01 | Antimicrobial [64] |
| Butyric acid | C4H8O2 | 153.36 ± 0.26 | Antimicrobial [65] |
| Carbamate | CH2NO2− | 0.92 ± 0.01 | Antimicrobial [66] |
| Carbamic acid | CH3NO2 | 1432.07 ± 4.62 | Antimicrobial [67] |
| Cephaloridine | C19H17N3O4S2 | 6.53 ± 0.01 | Antibiotic |
| Colchicine | C22H25NO6 | 15.06 ± 0.01 | Anti-inflammatory |
| Decanoic acid | C10H20O2 | 70.50 ± 0.01 | Antimicrobial [68] |
| Dihydroisosteviol | C20H32O3 | 5.44 ± 0.01 | Antimicrobial [69] |
| Docosahexaenoic acid | C22H32O2 | 5.79 ± 0.01 | Antimicrobial [70] |
| Docosanol | C22H46O | 6.41 ± 0.01 | Antimicrobial [71] |
| Dodecanamide | C12H25NO | 5.28 ± 0.01 | Antimicrobial [72] |
| Ethanol | C2H6O | 31.01 ± 0.30 | Antimicrobial [73] |
| Ethyl acetate | C4H8O2 | 44.86 ± 0.48 | Antimicrobial [74] |
| Galacturonic acid | C6H10O7 | 5.03 ± 0.05 | Antimicrobial [75] |
| D-gluco-hexodialdodifuranoside | C14H30O6 | 3.52 ± 0.01 | Anticancer |
| Gluconic acid | C6H12O7 | 307.05 ± 0.60 | Antimicrobial [76] |
| Glycerol | C3H8O3 | 336.23 ± 1.94 | Antimicrobial [77] |
| Glyoxylic acid | C2H2O3 | 4.68 ± 0.01 | Antimicrobial [78] |
| Griseoviridin | C22H27N3O7S | 3.14 ± 0.01 | Antibiotic |
| Guaiacol | C7H8O2 | 8.43 ± 0.01 | Antimicrobial [79] |
| Hexadecanoic acid | C16H32O2 | 147.85 ± 1.49 | Antimicrobial [80] |
| 2,4-Hexadienoic acid | C6H8O2 | 0.87 ± 0.01 | Antimicrobial [81] |
| Hexanedioic acid | C6H10O4 | 200.31 ± 0.01 | Antimicrobial [82] |
| Hexanoic acid | C6H12O2 | 20.24 ± 0.01 | Antimicrobial [83] |
| 2-Hydroxybenzaldehyde | C7H6O2 | 940.97 ± 1.64 | Antimicrobial [84] |
| 3-Hydroxybutyric acid | C4H8O3 | 18.19 ± 0.24 | Antimicrobial [85] |
| 4-Hydroxydiphenylamine | C12H11NO | 1.77 ± 0.01 | Antimicrobial [86] |
| 2-Hydroxyisocaproic acid | C6H12O3 | 173.95 ± 2.09 | Antimicrobial [87] |
| 3-(4-Hydroxyphenyl)propionic acid | C9H10O3 | 1.71 ± 0.01 | Anti-inflammatory |
| Indole | C8H7N | 22.32 ± 0.27 | Antimicrobial [88] |
| Indole-3-carboxylic acid | C9H7NO2 | 8.27 ± 0.01 | Antimicrobial [89] |
| Isocitric acid | C6H8O7 | 1.30 ± 0.01 | Antimicrobial [90] |
| 3-Isoxazolidinone | C3H5NO2 | 3.31 ± 0.01 | Antimicrobial [91] |
| Kaurene | C20H32 | 58.27 ± 0.10 | Antimicrobial [92] |
| 2-Keto-D-glucose | C6H10O6 | 2.11 ± 0.01 | Antibiotic |
| Lactic acid | C3H6O3 | 1055.38 ± 7.90 | Antimicrobial [93] |
| Linolenic acid | C18H30O2 | 8.00 ± 0.01 | Antimicrobial [94] |
| Lycopodine | C16H25NO | 4.13 ± 0.01 | Antimicrobial [95] |
| Malic acid | C4H6O5 | 21.02 ± 0.29 | Antimicrobial [96] |
| Mandelic acid | C8H8O3 | 443.50 ± 6.14 | Antimicrobial [97] |
| Meldrum’s acid | C6H8O4 | 1.49 ± 0.01 | Antimicrobial [98] |
| Methanol | CH4O | 10.15 ± 0.01 | Antimicrobial [99] |
| Methylenecyclopropane | C4H6 | 0.18 ± 0.01 | Antiviral [100] |
| Nonadecanoic acid | C19H38O2 | 2.48 ± 0.01 | Anticancer [101] |
| Nonanoic acid | C9H18O2 | 16.07 ± 0.01 | Antimicrobial [102] |
| Octadecanoic acid | C18H36O2 | 147.83 ± 1.66 | Antimicrobial [103] |
| Octanoic acid | C8H16O2 | 1.38 ± 0.01 | Antimicrobial [104] |
| Octenidine | C36H62N4 | 1.03 ± 0.01 | Antimicrobial [105] |
| Pentanedioic acid | C5H8O4 | 50.59 ± 0.64 | Antimicrobial [106] |
| Phenanthroline | C12H8N2 | 49.88 ± 0.39 | Antimicrobial [107] |
| 3-Phenyl-5-isoxazolone | C9H7NO2 | 2.14 ± 0.01 | Antimicrobial [108] |
| Phosphoric acid | H3PO4 | 0.68 ± 0.01 | Antimicrobial [109] |
| Propanamide | C3H7NO | 6.08 ± 0.01 | Antimicrobial [110] |
| Propanenitrile | C3H5N | 8.05 ± 0.01 | Antimicrobial [111] |
| Propanoic acid | C3H6O2 | 1184.32 ± 9.56 | Antimicrobial [112] |
| Propionamide | C3H7NO | 10.84 ± 0.01 | Antimicrobial [113] |
| Pteridine | C6H4N4 | 3.82 ± 0.03 | Antimicrobial [114] |
| Pyranone | C5H4O2 | 7.89 ± 0.07 | Antimicrobial [115] |
| Pyrazine | C4H4N2 | 33.19 ± 0.01 | Antimicrobial [116] |
| Pyridazine | C4H4N2 | 3.29 ± 0.01 | Antimicrobial [117] |
| Pyrrole | C4H5N | 3.04 ± 0.02 | Antimicrobial [118] |
| Pyrrolopyrimidine | C6H5N3 | 3.33 ± 0.01 | Antiviral [119] |
| Pyruvic acid | C3H4O3 | 56.20 ± 0.02 | Antimicrobial [120] |
| Quinazoline | C8H6N2 | 66.92 ± 0.01 | Antimicrobial [121] |
| Quinoline | C9H7N | 1.87 ± 0.01 | Antimicrobial [122] |
| 2-Quinolinone | C9H7NO | 284.36 ± 0.01 | Antimicrobial [123] |
| Sedoheptulose | C7H14O7 | 64.36 ± 0.01 | Antimicrobial [124] |
| Sesamol | C7H6O3 | 9.33 ± 0.07 | Antimicrobial [125] |
| Tartaric acid | C4H6O6 | 7.94 ± 0.01 | Antimicrobial [126] |
| Thiophene | C4H4S | 0.93 ± 0.01 | Antimicrobial [127] |
| Thiourea | CH4N2S | 2.16 ± 0.01 | Antimicrobial [128] |
| Thymol | C10H14O | 68.98 ± 0.01 | Antimicrobial [129] |
| 1,2,4-Triazole-3-carboxylic acid | C3H3N3O2 | 56.93 ± 0.01 | Antimicrobial [130] |
| Undecanoic acid | C11H22O2 | 2.09 ± 0.01 | Antimicrobial [131] |
| Urea | CH4N2O | 0.65 ± 0.01 | Antimicrobial [132] |
| Uridine | C9H12N2O6 | 1277.01 ± 3.34 | Antimicrobial [133] |
| Metabolite | Molecular Formula | Quantity (mg/L) | Biological Characteristic |
|---|---|---|---|
| Altronic acid | C6H12O7 | 0.22 ± 0.01 | Organic acid |
| Amphetamine | C9H13N | 1.35 ± 0.01 | Stimulant |
| Aromadendrene | C15H24 | 4.54 ± 0.01 | Essential oil |
| Benzene | C6H6 | 16.07 ± 0.01 | Aromatic |
| Benzocyclobutene | C8H8 | 7.98 ± 0.01 | Aromatic |
| Benzonitrile | C7H5N | 91.31 ± 0.01 | Aromatic |
| Butanal | C4H8O | 29.38 ± 0.19 | Aldehyde |
| Butane | C4H10 | 543.18 ± 5.86 | Alkane |
| Butanedioic acid | C4H6O4 | 31.11 ± 0.03 | Organic acid |
| Butanediol | C4H10O2 | 139.50 ± 0.03 | Alcohol |
| 1,2,2,3,4-Butanepentacarbonitrile | C9H5N5 | 0.55 ± 0.01 | Aromatic |
| Butanoic acid | C4H8O2 | 4399.87 ± 6.20 | Organic acid |
| 1-Butene | C4H8 | 379.99 ± 5.33 | Alkene |
| 1,4-Butenediol | C4H8O2 | 46.28 ± 0.01 | Alcohol |
| 2-Butenoic acid | C4H6O2 | 52.41 ± 0.11 | Organic acid |
| 3-Buten-1-ol | C4H8O | 2.31 ± 0.01 | Alcohol |
| Butylamine | C4H11N | 84.67 ± 0.01 | Amine |
| Butyne | C4H6 | 7.46 ± 0.01 | Alkyne |
| Butynol | C4H10O | 0.43 ± 0.01 | Alcohol |
| Butyrate | C4H7O2− | 0.72 ± 0.01 | Flavoring |
| Camphoric acid | C10H16O4 | 0.81 ± 0.01 | Organic acid |
| Carbophenoxon sulfone | C11H16ClO5PS2 | 11.46 ± 0.01 | Organosulfone |
| Cholestane | C27H48 | 3.63 ± 0.01 | Cholesterol |
| 1-Cholestene | C27H46 | 31.64 ± 0.01 | Cholesterol |
| Cholestenone | C27H44O | 7.61 ± 0.09 | Cholesterol |
| Cholesterol | C27H46O | 75.80 ± 0.66 | Cholesterol |
| Chromium | Cr | 2.53 ± 0.01 | Mineral |
| Cortisone | C21H28O5 | 2.09 ± 0.01 | Steroid |
| Cyclobutanemethanol | C5H10O | 25.00 ± 0.01 | Aromatic |
| Cyclohexane | C6H12 | 4.64 ± 0.03 | Aromatic |
| Cyclohexene | C6H10 | 1.31 ± 0.01 | Aromatic |
| 3-Cyclohexene-1-methanol | C7H12O | 2.78 ± 0.01 | Essential oil |
| 1-Cyclohexyl-tetradecane | C20H40 | 5.00 ± 0.01 | Aromatic |
| Cyclopenta[de]naphthalene | C12H8 | 8.26 ± 0.01 | Aromatic |
| Cyclopentane | C5H10 | 4.84 ± 0.01 | Aromatic |
| 1,2,4-Cyclopentanetrione | C5H4O3 | 7.93 ± 0.01 | Aromatic |
| Cyclopentene | C5H8 | 0.61 ± 0.01 | Aromatic |
| Cyclopropanecarboxylic acid | C4H6O2 | 3.04 ± 0.01 | Organic acid |
| Decane | C10H22 | 1.98 ± 0.01 | Alkane |
| 1-Decanol | C10H22O | 1.75 ± 0.01 | Fatty alcohol |
| 2,6-Diamino-4-hexynoic acid | C6H10N2O2 | 2.10 ± 0.01 | Organic acid |
| 1,3-Diazepane-2,4,6-trione | C5H6N2O3 | 1.09 ± 0.01 | Aromatic |
| 3-Dibenzofuranamine | C12H9NO | 19.35 ± 0.18 | Aromatic |
| Diethylene glycol | C4H10O3 | 67.89 ± 0.68 | Solvent |
| 2,3-Dihydroxybutanoic acid | C4H8O4 | 13.75 ± 0.07 | Organic acid |
| 1,1-Diisobutoxybutane | C12H26O2 | 1.72 ± 0.01 | Aldehyde |
| Diisopropyl malonate | C9H16O4 | 1.30 ± 0.01 | Acid ester |
| Dimethylbutanedioate | C6H10O4 | 0.77 ± 0.01 | Flavoring |
| 2,3-Dimethylbutanoic acid | C6H12O2 | 2129.98 ± 3.01 | Fatty acid |
| 3,3-Dimethyl-1-butanol | C6H14O | 18.60 ± 0.01 | Alcohol |
| Dimethylcyclohexanone | C8H14O | 89.00 ± 0.01 | Aromatic |
| Dimethyldecahydronaphthalene | C12H22 | 56.95 ± 0.01 | Aromatic |
| Dimethyl malonate | C5H8O4 | 8.30 ± 0.01 | Acid ester |
| Dipropylacetic acid | C8H16O2 | 120.83 ± 0.01 | Organic acid |
| 13,16-Docasadienoic acid | C22H40O2 | 51.71 ± 0.35 | Fatty acid |
| Docosanoic acid | C22H44O2 | 2.40 ± 0.01 | Fatty acid |
| 13-Docosenamide | C22H43NO | 64.20 ± 0.78 | Fatty amide |
| Dodecane | C12H26 | 1462.61 ± 0.01 | Alkane |
| Dodecanedioic acid | C12H22O4 | 4.84 ± 0.01 | Organic acid |
| 5,8,11-Eicosatriynoic acid | C20H28O2 | 0.97 ± 0.01 | Fatty acid |
| Estratetraenol | C18H22O | 15.35 ± 0.01 | Steroid |
| Ethane | C2H6 | 37.25 ± 0.01 | Alkane |
| Ethanedioic acid | C2H2O4 | 16.61 ± 0.19 | Organic acid |
| Ethanesulfonic acid | C2H6O3S | 3.13 ± 0.02 | Sulfonic acid |
| Ethanimidic acid | C4H9NO | 0.63 ± 0.01 | Organic acid |
| Ethyl butyrate | C6H12O2 | 7.25 ± 0.02 | Flavoring |
| Ethylene | C2H4 | 1.99 ± 0.01 | Alkene |
| Ethylene glycol | C2H6O2 | 141.94 ± 0.01 | Solvent |
| 3-Furanacetaldehyde | C6H6O2 | 0.75 ± 0.01 | Aldehyde |
| 2-Furancarboxylic acid | C5H4O3 | 1.98 ± 0.01 | Organic acid |
| 2-Furanone | C4H4O2 | 3.41 ± 0.02 | Flavoring |
| Glucuronolactone | C6H8O6 | 3.95 ± 0.04 | Lactone |
| Glyceraldehyde acetonide | C6H10O3 | 247.79 ± 0.01 | Carboxaldehyde |
| L-gulono-1,4-lactone | C6H10O6 | 21.25 ± 0.23 | Lactone |
| Heptadecane | C17H36 | 7.06 ± 0.01 | Alkane |
| Heptadecane-1,2-diol | C17H36O2 | 54.71 ± 0.01 | Fatty alcohol |
| 3-Heptyn-1-ol | C7H12O | 10.73 ± 0.01 | Fatty alcohol |
| Heptanamide | C7H15NO | 14.97 ± 0.01 | Fatty amide |
| Hexadecanamide | C16H33NO | 5.81 ± 0.01 | Fatty amide |
| Hexadecane | C16H34 | 97.90 ± 0.01 | Alkane |
| 1-Hexene | C6H12 | 10.50 ± 0.01 | Alkene |
| 3-Hexenedioic acid | C6H8O4 | 11.76 ± 0.01 | Fatty acid |
| 3-Hexen-1-ol | C6H12O | 3.00 ± 0.01 | Fatty alcohol |
| 4-Hexen-1-yne | C6H8 | 2.27 ± 0.01 | Alkyne |
| 3-Hydroxy-2-butanone | C4H8O2 | 72.50 ± 0.01 | Methyl ketone |
| 2-Hydroxyglutaric acid | C5H8O5 | 2.30 ± 0.01 | Organic acid |
| 3-Hydroxypyruvic acid | C3H4O4 | 1.32 ± 0.01 | Organic acid |
| 3-Hydroxysebacic acid | C10H18O5 | 4.09 ± 0.03 | Organic acid |
| Inabenfide | C19H15ClN2O2 | 3.19 ± 0.01 | Herbicide |
| Iron | Fe | 3.25 ± 0.01 | Mineral |
| 2-Ketobutyric acid | C4H6O3 | 222.57 ± 0.02 | Organic acid |
| 2-Ketohexanoic acid | C6H10O3 | 1.03 ± 0.01 | Fatty acid |
| Ketovaleric acid | C5H8O3 | 17.59 ± 0.01 | Ketoacid |
| Malonic acid | C3H4O4 | 0.25 ± 0.01 | Organic acid |
| Methanaminium | CH6N | 5.93 ± 0.05 | Conjugate acid |
| Methyl butyrate | C5H10O2 | 9.83 ± 0.01 | Flavoring |
| Methylcyclopentadiene | C6H8 | 465.49 ± 0.01 | Aromatic |
| 6-Methyl-3,5-heptadien-2-one | C8H12O | 106.73 ± 0.48 | Flavoring |
| Methyl phenyl sulfoxide | C7H8OS | 2.38 ± 0.01 | Aromatic |
| 2-Methylpropanoic acid | C4H8O2 | 8.21 ± 0.01 | Organic acid |
| 2-Methylpropene | C4H8 | 69.91 ± 0.31 | Alkene |
| 2-Methyl-4-propyl-1,3-oxathiane | C8H16OS | 53.44 ± 0.01 | Flavoring |
| Methyl tetradecanoate | C15H30O2 | 3.53 ± 0.01 | Flavoring |
| 4-Methyl-5-thiazoleethanol | C6H9NOS | 120.92 ± 0.01 | Flavoring |
| Methyl valerate | C6H12O2 | 445.57 ± 0.01 | Flavoring |
| 3-Methylvaleric acid | C6H12O2 | 5.55 ± 0.01 | Fatty acid |
| Monoethyl malonic acid | C5H8O4 | 13.41 ± 0.16 | Organic acid |
| Monostearin | C21H42O4 | 35.76 ± 0.31 | Emulsifier |
| Morphine | C17H19NO3 | 7.05 ± 0.01 | Painkiller |
| N-acetyl-glucosamine | C8H15NO6 | 24.93 ± 0.33 | Chitosan |
| Nickel | Ni | 6.56 ± 0.01 | Mineral |
| Nonane | C9H20 | 2.28 ± 0.01 | Alkane |
| 5-Norbornene-2-carboxylic acid | C8H10O2 | 0.93 ± 0.01 | Organic acid |
| Octadecanamide | C18H37NO | 47.22 ± 0.01 | Fatty amide |
| Octadecane | C18H38 | 137.50 ± 0.01 | Alkane |
| Octadecenamide | C18H35NO | 67.10 ± 0.77 | Fatty amide |
| 17-Octadecynoic acid | C18H32O2 | 18.04 ± 0.01 | Fatty acid |
| Octahydronaphthalene | C10H16 | 3.48 ± 0.01 | Aromatic |
| Octahydronaphthalene-1,4-diol | C10H16O2 | 57.23 ± 0.01 | Alcohol |
| γ-Octalactone | C8H14O2 | 238.91 ± 0.37 | Flavoring |
| Octane | C8H18 | 25.30 ± 0.01 | Alkane |
| 1-Octene | C8H16 | 10.56 ± 0.01 | Alkene |
| Oleic acid | C18H34O2 | 17.59 ± 0.01 | Fatty acid |
| 3-Oxooctanoic acid | C8H14O3 | 1185.00 ± 0.01 | Fatty acid |
| 2-Oxovaleric acid | C5H8O3 | 4.12 ± 0.01 | Ketoacid |
| Para-methoxy-N-methylamphetamine | C11H17NO | 145.28 ± 0.01 | Stimulant |
| Pentadecanoic acid | C15H30O2 | 11.42 ± 0.01 | Fatty acid |
| Pentaethylene glycol | C22H46O6 | 6.10 ± 0.01 | Solvent |
| Pentadecane | C15H32 | 2.32 ± 0.01 | Alkane |
| Pentanamide | C5H11NO | 1.41 ± 0.01 | Acid amide |
| Pentane | C5H12 | 7.22 ± 0.09 | Alkane |
| Pentanoic acid | C5H10O2 | 26.12 ± 0.20 | Flavoring |
| Pentaoxacyclopentadecane | C10H20O5 | 1.60 ± 0.01 | Crown ether |
| Pentenedioate | C5H6O42− | 2.88 ± 0.01 | Organic acid |
| Pentenedioic acid | C5H6O4 | 7.41 ± 0.07 | Organic acid |
| 2-Pentenoic acid | C5H8O2 | 70.15 ± 0.95 | Organic acid |
| 9-O-pivaloyl-N-acetylcolchinol | C25H31NO6 | 24.44 ± 0.17 | Aromatic |
| Pregnenolone | C21H32O2 | 3.80 ± 0.01 | Steroid |
| Propanal | C3H6O | 2.77 ± 0.02 | Aldehyde |
| Propane | C3H8 | 6.39 ± 0.02 | Alkane |
| Propanedioic acid | C3H4O4 | 10.46 ± 0.01 | Organic acid |
| 1,3-Propanediol | C3H8O2 | 3.03 ± 0.01 | Alcohol |
| 1,2,3-Propanetriol | C3H8O3 | 1595.72 ± 0.02 | Polyol |
| Propanone | C3H6O | 114.26 ± 1.34 | Ketone |
| 2-Propenamide | C3H5NO | 338.39 ± 4.16 | Fatty amide |
| 2-Propenoic acid | C3H4O2 | 15.90 ± 0.01 | Organic acid |
| Propylamine | C3H9N | 3.25 ± 0.01 | Fatty amine |
| Propylene glycol | C3H8O2 | 2458.09 ± 1.26 | Solvent |
| Pseudoephedrine | C10H15NO | 24.34 ± 0.01 | Decongestant |
| Pseudouridine | C9H12N2O6 | 2.99 ± 0.01 | Nucleoside |
| Pyrandiol | C5H6O3 | 40.72 ± 0.01 | Alcohol |
| Pyruvate oxime | C3H5NO3 | 14.13 ± 0.09 | Acid amine |
| Scopolin | C16H18O9 | 179.76 ± 2.08 | Phytochemical |
| Sebacic acid | C10H18O4 | 3.53 ± 0.01 | Fatty acid |
| Succinate | C4H4O42- | 0.78 ± 0.01 | Flavoring |
| Succinonitrile | C4H4N2 | 0.16 ± 0.01 | Nitrile |
| Talonic acid | C6H12O7 | 5.55 ± 0.01 | Organic acid |
| Tetradecane | C14H30 | 14.12 ± 0.01 | Alkane |
| Tetradecanoic acid | C14H28O2 | 7.86 ± 0.03 | Fatty acid |
| 1-Tetradecanol | C14H30O | 1.24 ± 0.01 | Fatty alcohol |
| Tetraethylene glycol | C8H18O5 | 0.53 ± 0.01 | Solvent |
| 1,2,4,5-Tetramethylbenzene | C10H14 | 650.91 ± 0.01 | Aromatic |
| Thiodiglycol | C4H10O2S | 12.39 ± 0.14 | Alcohol |
| Tricyclodecenyl propionate | C13H18O2 | 2.31 ± 0.01 | Fragrance |
| Tridecane | C13H28 | 207.98 ± 0.01 | Alkane |
| Tridecanoic acid | C13H26O2 | 15.33 ± 0.01 | Fatty acid |
| Triethylene glycol | C6H14O4 | 120.91 ± 0.04 | Solvent |
| 2,3,4-Trihydroxybutanoic acid | C4H8O5 | 41.02 ± 0.31 | Organic acid |
| 2,4,5-Trihydroxypentanoic acid | C5H10O5 | 18.61 ± 0.01 | Organic acid |
| 1,2,4-Trimethylbenzene | C9H12 | 16.28 ± 0.01 | Aromatic |
| 1-Undecene | C11H22 | 7.66 ± 0.03 | Alkene |
| Vitamin C | C6H8O6 | 9.22 ± 0.01 | Ascorbic acid |
| Metabolite | Molecular Formula | Quantity (mg/L) | Biological Characteristic |
|---|---|---|---|
| Altro-heptulose | C7H14O7 | 6.79 ± 0.01 | Sugar substitute |
| Arabinitol | C5H15O5 | 115.06 ± 0.03 | Sugar alcohol |
| Arabinofuranose | C5H10O5 | 1714.08 ± 2.23 | Sugar substitute |
| Arabinofuranoside | C5H9O5 | 592.25 ± 0.06 | Sugar substitute |
| D-arabino-3-hexulose | C6H12O6 | 17.21 ± 0.01 | Sugar substitute |
| Arabinonic acid | C5H10O6 | 81.43 ± 1.14 | Sugar acid |
| Arabinopyranose | C5H10O5 | 2350.31 ± 1.98 | Sugar substitute |
| Arabinose | C5H10O5 | 912.65 ± 2.11 | Sugar substitute |
| Arabitol | C5H10O5 | 1456.58 ± 0.56 | Sugar alcohol |
| 3-Deoxy-D-arabino-hexonic acid | C6H12O6 | 112.61 ± 0.11 | Sugar acid |
| 2-Deoxy-erythro-pentofuranose | C5H10O4 | 596.47 ± 8.29 | Sugar substitute |
| 3-Deoxy-erythro-pentonic acid | C5H10O5 | 43.20 ± 0.03 | Sugar acid |
| 2-Deoxy-erythro-pentopyranose | C5H10O4 | 2.50 ± 0.01 | Sugar substitute |
| 2-Deoxy-erythro-pentose | C5H10O4 | 42.49 ± 0.36 | Sugar substitute |
| 2-Deoxy-D-galactopyranose | C6H12O5 | 281.36 ± 2.07 | Sugar substitute |
| 2-Deoxy-D-glucose | C6H12O5 | 677.13 ± 0.01 | Sugar substitute |
| Deoxy-ribose | C5H10O4 | 19.08 ± 0.01 | Sugar substitute |
| 3-Deoxy-D-ribohexonic acid | C6H12O | 20.71 ± 0.01 | Sugar acid |
| Dihydroxyacetone | C3H6O3 | 21.99 ± 0.01 | Sugar substitute |
| Dulcitol | C6H14O6 | 3323.48 ± 4.53 | Sugar alcohol |
| Erythritol | C4H10O4 | 0.26 ± 0.01 | Sugar alcohol |
| Erythro-pentitol | C5H12O | 29.50 ± 0.03 | Sugar alcohol |
| Erythrose | C4H8O4 | 522.45 ± 5.06 | Sugar substitute |
| Erythro-tetrofuranose | C5H10O5 | 22.65 ± 0.01 | Sugar substitute |
| Fructopyranose | C6H12O6 | 6.94 ± 0.01 | Sugar substitute |
| Fructose | C6H12O6 | 2462.43 ± 1.42 | Sugar substitute |
| Fructose oxime | C6H13NO6 | 2421.99 ± 0.01 | Sugar substitute |
| Galactofuranose | C6H12O6 | 1076.99 ± 0.78 | Sugar substitute |
| Galactoheptulose | C7H14O7 | 2.20 ± 0.01 | Sugar substitute |
| Galactopyranose | C6H12O6 | 943.28 ± 2.36 | Sugar substitute |
| Galactose | C6H12O6 | 8909.55 ± 1.18 | Sugar substitute |
| Galactose oxime | C6H13NO6 | 369.21 ± 0.39 | Sugar substitute |
| Glucaric acid | C6H10O8 | 0.08 ± 0.01 | Sugar acid |
| Glucitol | C6H14O6 | 601.89 ± 7.85 | Sugar alcohol |
| Glucofuranose | C6H12O6 | 2035.95 ± 1.10 | Sugar substitute |
| Glucopyranose | C6H12O6 | 7024.77 ± 8.40 | Sugar substitute |
| Glucopyranuronic acid | C6H10O7 | 1239.42 ± 0.13 | Sugar acid |
| Glucose | C6H12O6 | 2010.62 ± 1.31 | Sugar substitute |
| Glucose oxime | C6H13NO6 | 610.91 ± 0.23 | Sugar substitute |
| Glucuronic acid | C6H10O7 | 7.58 ± 0.07 | Sugar acid |
| Glutaconic acid | C5H6O4 | 0.77 ± 0.01 | Sugar acid |
| Glyceraldehyde | C3H6O3 | 422.51 ± 1.30 | Sugar substitute |
| D-glycero-D-galacto-heptose | C7H14O7 | 91.04 ± 0.01 | Sugar substitute |
| D-glycero-D-gluco-heptose | C7H14O7 | 129.78 ± 1.66 | Sugar substitute |
| D-glycero-D-gulo-heptonic acid | C7H14O8 | 40.26 ± 0.14 | Sugar acid |
| D-glycero-L-manno-heptonic acid | C7H14O8 | 1192.77 ± 2.99 | Sugar acid |
| Gulonic acid | C6H12O7 | 14.17 ± 0.10 | Sugar acid |
| Gulose | C6H12O6 | 361.01 ± 2.59 | Sugar substitute |
| Lactose | C12H22O11 | 125.72 ± 0.01 | Sugar substitute |
| Levoglucosan | C6H10O5 | 1.32 ± 0.01 | Sugar substitute |
| Lyxopyranose | C5H10O5 | 1654.75 ± 1.82 | Sugar substitute |
| Lyxose | C5H10O5 | 603.81 ± 2.35 | Sugar substitute |
| Maltose | C12H22O11 | 5653.05 ± 5.87 | Sugar substitute |
| Mannitol | C6H14O6 | 177.42 ± 0.80 | Sugar alcohol |
| Mannofuranose | C6H12O6 | 1009.76 ± 5.57 | Sugar substitute |
| Mannofuranuronic acid | C6H8O6 | 51.35 ± 0.01 | Sugar acid |
| Mannonic acid | C6H12O7 | 2567.14 ± 1.67 | Sugar acid |
| Mannopyranose | C6H12O | 358.11 ± 0.21 | Sugar substitute |
| Mannose | C6H12O6 | 5744.85 ± 7.73 | Sugar substitute |
| Melibiose | C12H22O11 | 0.69 ± 0.01 | Sugar substitute |
| 2,5-Methylene-D,L-rhamnitol | C7H14O5 | 1.33 ± 0.01 | Sugar substitute |
| Methyl-D-galactofuranoside | C7H14O6 | 481.33 ± 1.13 | Sugar substitute |
| Methyl-D-glucopyranoside | C7H14O6 | 4908.57 ± 6.54 | Sugar substitute |
| Methyl-D-lyxofuranoside | C6H12O5 | 499.24 ± 4.75 | Sugar substitute |
| Methyl-D-mannopyranoside | C7H14O6 | 131.27 ± 0.01 | Sugar substitute |
| Methyl-D-ribofuranoside | C6H12O | 120.28 ± 0.01 | Sugar substitute |
| Methyl-D-xylopyranoside | C6H12O5 | 1.23 ± 0.01 | Sugar substitute |
| Myo-inositol | C6H12O6 | 130.33 ± 0.50 | Sugar substitute |
| Pentitol | C5H12O5 | 185.84 ± 0.01 | Sugar Alcohol |
| Phenyl-D-galactopyranoside | C12H16O6 | 9584.87 ± 0.01 | Sugar substitute |
| D-ribo-2-hexulose | C6H12O6 | 5.77 ± 0.03 | Sugar substitute |
| Ribonic acid | C5H10O6 | 89.50 ± 0.70 | Sugar acid |
| Ribopyranose | C5H10O5 | 1427.89 ± 7.29 | Sugar substitute |
| Ribose | C5H10O5 | 249.45 ± 3.50 | Sugar substitute |
| Sorbopyranose | C6H12O6 | 39.12 ± 0.11 | Sugar substitute |
| Talose | C6H12O6 | 800.65 ± 2.15 | Sugar substitute |
| Threitol | C4H10O4 | 121.12 ± 1.66 | Sugar alcohol |
| Threonic acid | C4H8O5 | 30.15 ± 0.01 | Sugar acid |
| Turanose | C12H22O11 | 66.04 ± 0.40 | Sugar substitute |
| Xylitol | C5H12O5 | 508.27 ± 0.44 | Sugar alcohol |
| Xylofuranose | C5H10O5 | 88.69 ± 0.01 | Sugar substitute |
| D-xylo-hexulose | C6H12O6 | 2.66 ± 0.02 | Sugar substitute |
| Xylonic acid | C5H10O6 | 156.31 ± 1.43 | Sugar acid |
| Xylofuranose | C5H10O5 | 88.69 ± 0.01 | Sugar substitute |
| Xylopyranose | C5H10O5 | 915.86 ± 0.30 | Sugar substitute |
| Xylose | C5H10O5 | 1417.45 ± 1.36 | Sugar substitute |
| Xylulose | C5H10O5 | 7.30 ± 0.01 | Sugar substitute |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Y.N.; Zhang, J.H.; Chen, W.N. GC-MS-Based Metabolomics Analysis of Prawn Shell Waste Co-Fermentation by Lactobacillus plantarum and Bacillus subtilis. Polysaccharides 2020, 1, 31-50. https://doi.org/10.3390/polysaccharides1010004
Tan YN, Zhang JH, Chen WN. GC-MS-Based Metabolomics Analysis of Prawn Shell Waste Co-Fermentation by Lactobacillus plantarum and Bacillus subtilis. Polysaccharides. 2020; 1(1):31-50. https://doi.org/10.3390/polysaccharides1010004
Chicago/Turabian StyleTan, Yun Nian, Jian Hua Zhang, and Wei Ning Chen. 2020. "GC-MS-Based Metabolomics Analysis of Prawn Shell Waste Co-Fermentation by Lactobacillus plantarum and Bacillus subtilis" Polysaccharides 1, no. 1: 31-50. https://doi.org/10.3390/polysaccharides1010004
APA StyleTan, Y. N., Zhang, J. H., & Chen, W. N. (2020). GC-MS-Based Metabolomics Analysis of Prawn Shell Waste Co-Fermentation by Lactobacillus plantarum and Bacillus subtilis. Polysaccharides, 1(1), 31-50. https://doi.org/10.3390/polysaccharides1010004





