High-Fat-High-Fructose Diet Elicits Brown Adipocyte Dysfunction through miRNA-103 Induced miRNA Biogenesis Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. mRNA and miRNA Extraction and RT-PCR
2.3. Western Blot Analysis
2.4. Transient Transfections and miRNA-103 Knockdown Assay
2.5. Bioinformatic Analysis
2.6. Statistical Analysis
3. Results
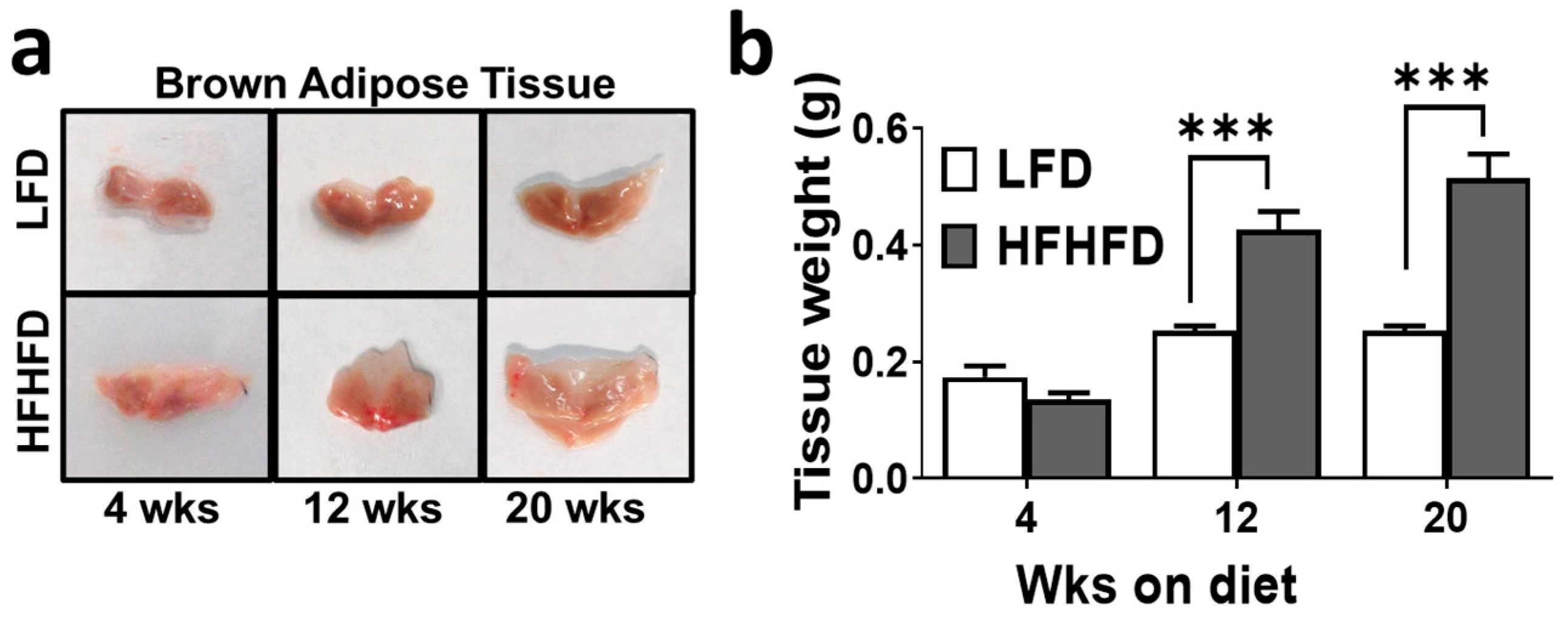
3.1. HFHFD-Fed Mice Exhibit Enlarged and Whitened BAT
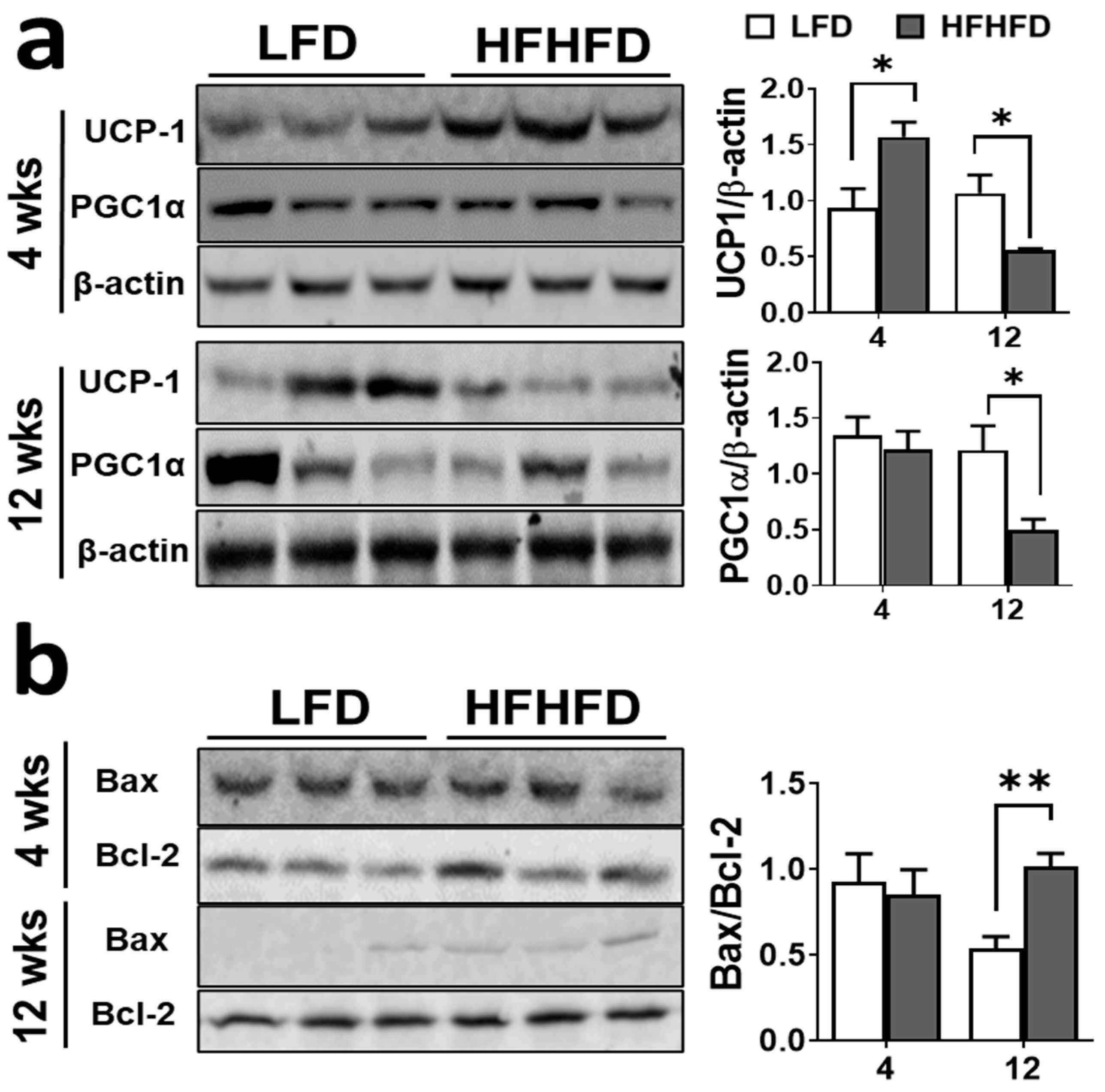
3.2. HFHFD Feeding Impairs BAT Function
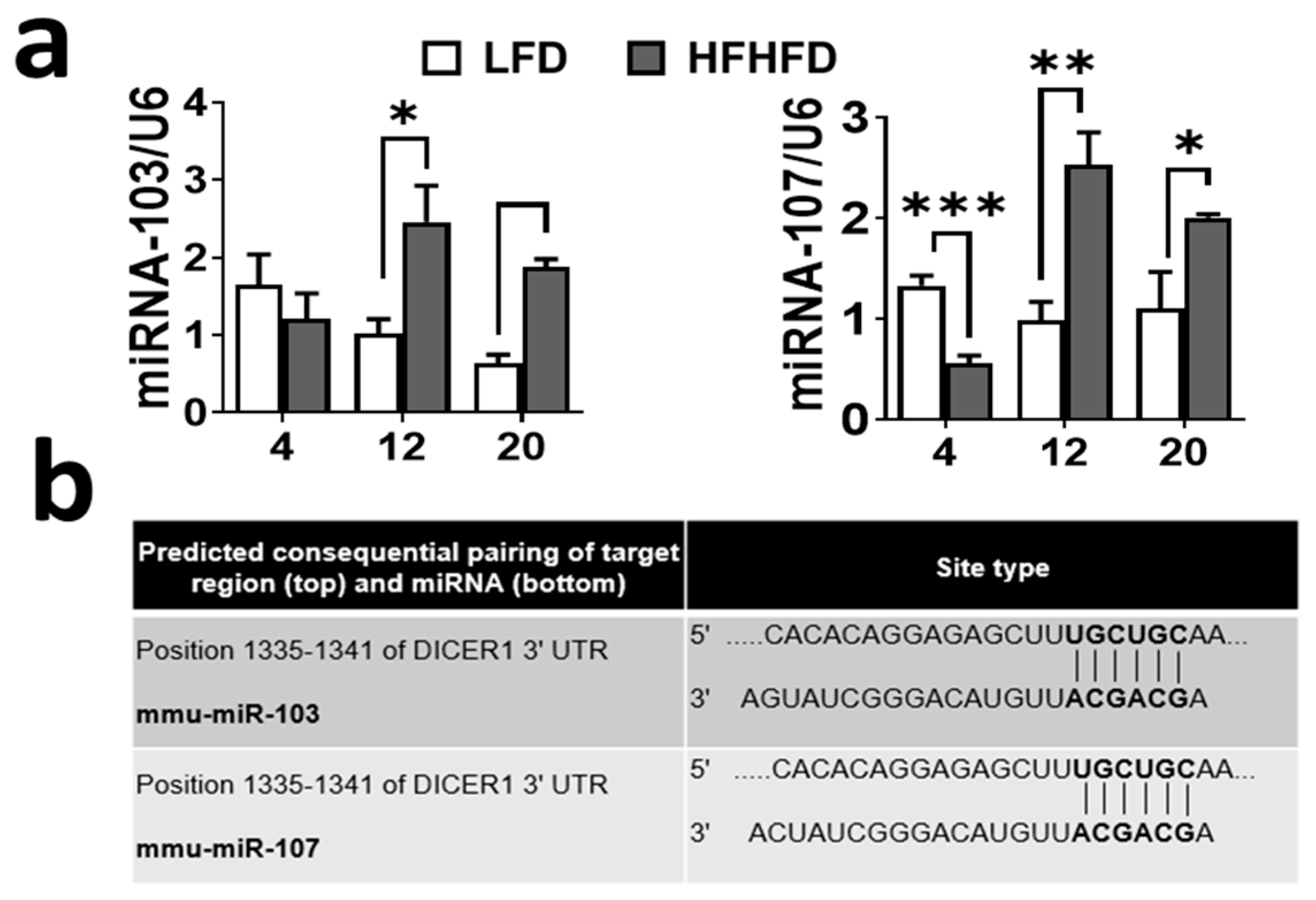
3.3. Upregulation of miRNA-103/107 Expression in HFHFD-Fed Mice
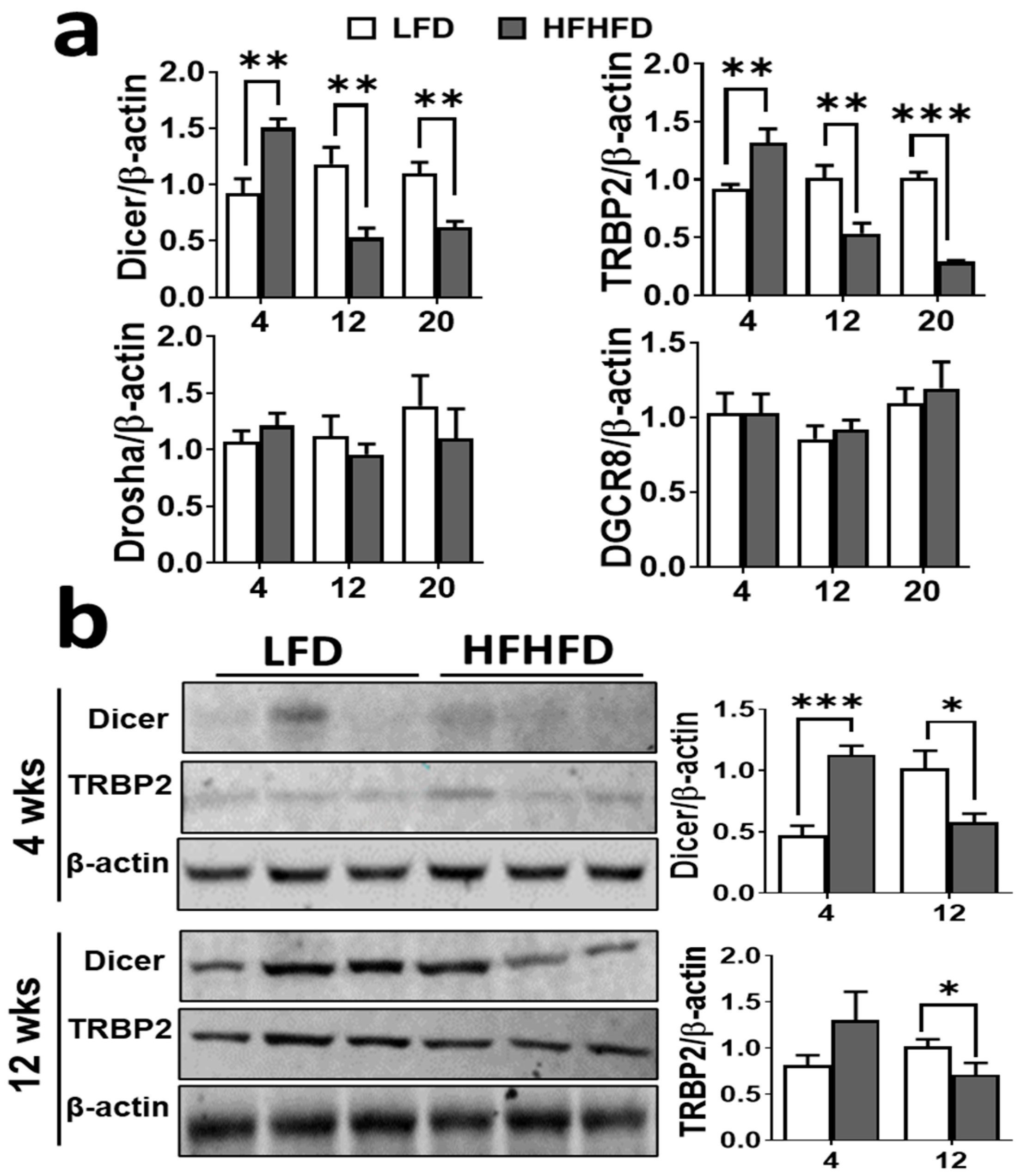
3.4. HFHFD Feeding Inhibits miRNA Biogenesis Machinery
3.5. miRNA-103 Mediated Regulation of miRNA Biogenesis Machinery In Vitro
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Targets | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Dicer1 | GGTCCTTTCTTTGGACTGCCA | GCGATGAACGTCTTCCCTGA |
| TRBP2 | AGGAGCAGGCTTTCCATGTC | GGTGGACAGTTCCACTAGGC |
| Drosha | ATGCAAGGCAATACGTGTCAT | TTTTGGGGTCTGAAAGCTGGT |
| DGCR8 | GCAGGAGAAGCGATGATGGAG | CCGTAGAAGTTGAATGGGTCG |
| UCP1 | ACTGCCACACCTCCAGTCATT | CTTTGCCTCACTCAGGATTGG |
| PGC1α | TATGGAGTGACATAGAGTGTGCT | CCACTTCAATCCACCCAGAAAG |
| β-actin | AGCCATGTACGTAGCCATCC | GCTGTGGTGGTGAAGCTGTA |
| miRNA-103-3p | GCAGAGCAGCATTGTACAG | GGTCCAGTTTTTTTTTTTTTTTCATAG |
| miRNA-107-3p | GCAGAGCAGCATTGTACAG | GGTCCAGTTTTTTTTTTTTTTTGATAG |
| U6 | TGGCCCCTGCGCAAGGATG | AGTTTTTTTTTTTTTTTGCGCAG |
References
- Mitchell, N.S.; Catenacci, V.A.; Wyatt, H.R.; Hill, J.O. Obesity: Overview of an epidemic. Psychiatr. Clin. N. Am. 2011, 34, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Crowley, V.E.; Yeo, G.S.; O’Rahilly, S. Obesity therapy: Altering the energy intake-and-expenditure balance sheet. Nat. Rev. Drug Discov. 2002, 1, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.B.; Srivastava, G.; Reid, T.J.; Aronne, L.J. Understanding the pathophysiologic pathways that underlie obesity and options for treatment. Expert. Rev. Endocrinol. Metab. 2021, 16, 321–338. [Google Scholar] [CrossRef] [PubMed]
- Naomi, R.; Teoh, S.H.; Halim, S.; Embong, H.; Hasain, Z.; Bahari, H.; Kumar, J. Unraveling Obesity: Transgenerational Inheritance, Treatment Side Effects, Flavonoids, Mechanisms, Microbiota, Redox Balance, and Bioavailability—A Narrative Review. Antioxidants 2023, 12, 1549. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.Y.; Primus Dass, K.T.; Tsai, S.F.; Chuang, H.M.; Lin, S.Z.; Liu, S.P.; Harn, H.J. Clinical Application Potential of Small Molecules that Induce Brown Adipose Tissue Thermogenesis by Improving Fat Metabolism. Cell Transplant. 2020, 29, 963689720927394. [Google Scholar] [CrossRef] [PubMed]
- Lowell, B.B.; S-Susulic, V.; Hamann, A.; Lawitts, J.A.; Himms-Hagen, J.; Boyer, B.B.; Kozak, L.P.; Flier, J.S. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 1993, 366, 740–742. [Google Scholar] [CrossRef] [PubMed]
- Becher, T.; Palanisamy, S.; Kramer, D.J.; Eljalby, M.; Marx, S.J.; Wibmer, A.G.; Butler, S.D.; Jiang, C.S.; Vaughan, R.; Schöder, H.; et al. Brown adipose tissue is associated with cardiometabolic health. Nat. Med. 2021, 27, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Herz, C.T.; Kulterer, O.C.; Prager, M.; Schmöltzer, C.; Langer, F.B.; Prager, G.; Marculescu, R.; Kautzky-Willer, A.; Hacker, M.; Haug, A.R.; et al. Active Brown Adipose Tissue is Associated with a Healthier Metabolic Phenotype in Obesity. Diabetes 2021, 71, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Frontini, A.; Cinti, S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab. 2010, 11, 253–256. [Google Scholar] [CrossRef]
- Heenan, K.A.; Carrillo, A.E.; Fulton, J.L.; Ryan, E.J.; Edsall, J.R.; Rigopoulos, D.; Markofski, M.M.; Flouris, A.D.; Dinas, P.C. Effects of Nutrition/Diet on Brown Adipose Tissue in Humans: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 2752. [Google Scholar] [CrossRef]
- Feldmann, H.M.; Golozoubova, V.; Cannon, B.; Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009, 9, 203–209. [Google Scholar] [CrossRef]
- Srivastava, S.; Baxa, U.; Niu, G.; Chen, X.; Veech, R.L. A ketogenic diet increases brown adipose tissue mitochondrial proteins and UCP1 levels in mice. IUBMB Life 2013, 65, 58–66. [Google Scholar] [CrossRef]
- Roberts-Toler, C.; O’Neill, B.T.; Cypess, A.M. Diet-induced obesity causes insulin resistance in mouse brown adipose tissue. Obesity 2015, 23, 1765–1770. [Google Scholar] [CrossRef]
- Shimizu, I.; Aprahamian, T.; Kikuchi, R.; Shimizu, A.; Papanicolaou, K.N.; MacLauchlan, S.; Maruyama, S.; Walsh, K. Vascular rarefaction mediates whitening of brown fat in obesity. J. Clin. Investig. 2014, 124, 2099–2112. [Google Scholar] [CrossRef] [PubMed]
- Alcala, M.; Calderon-Dominguez, M.; Bustos, E.; Ramos, P.; Casals, N.; Serra, D.; Viana, M.; Herrero, L. Increased inflammation, oxidative stress and mitochondrial respiration in brown adipose tissue from obese mice. Sci. Rep. 2017, 7, 16082. [Google Scholar] [CrossRef]
- Kuipers, E.N.; Held, N.M.; In Het Panhuis, W.; Modder, M.; Ruppert, P.M.M.; Kersten, S.; Kooijman, S.; Guigas, B.; Houtkooper, R.H.; Rensen, P.C.N.; et al. A single day of high-fat diet feeding induces lipid accumulation and insulin resistance in brown adipose tissue in mice. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E820–E830. [Google Scholar] [CrossRef]
- Richard, G.; Blondin, D.P.; Syed, S.A.; Rossi, L.; Fontes, M.E.; Fortin, M.; Phoenix, S.; Frisch, F.; Dubreuil, S.; Guérin, B.; et al. High-fructose feeding suppresses cold-stimulated brown adipose tissue glucose uptake independently of changes in thermogenesis and the gut microbiome. Cell Rep. Med. 2022, 3, 100742. [Google Scholar] [CrossRef] [PubMed]
- Raad, G.; Serra, F.; Martin, L.; Derieppe, M.A.; Gilleron, J.; Costa, V.L.; Pisani, D.F.; Amri, E.Z.; Trabucchi, M.; Grandjean, V. Paternal multigenerational exposure to an obesogenic diet drives epigenetic predisposition to metabolic diseases in mice. eLife 2021, 10, e61736. [Google Scholar] [CrossRef]
- Martino, E.; D’Onofrio, N.; Balestrieri, A.; Colloca, A.; Anastasio, C.; Sardu, C.; Marfella, R.; Campanile, G.; Balestrieri, M.L. Dietary Epigenetic Modulators: Unravelling the Still-Controversial Benefits of miRNAs in Nutrition and Disease. Nutrients 2024, 16, 160. [Google Scholar] [CrossRef]
- MacDonald-Ramos, K.; Martínez-Ibarra, A.; Monroy, A.; Miranda-Ríos, J.; Cerbón, M. Effect of Dietary Fatty Acids on MicroRNA Expression Related to Metabolic Disorders and Inflammation in Human and Animal Trials. Nutrients 2021, 13, 1830. [Google Scholar] [CrossRef]
- Quintanilha, B.J.; Pinto Ferreira, L.R.; Ferreira, F.M.; Neto, E.C.; Sampaio, G.R.; Rogero, M.M. Circulating plasma microRNAs dysregulation and metabolic endotoxemia induced by a high-fat high-saturated diet. Clin. Nutr. 2020, 39, 554–562. [Google Scholar] [CrossRef]
- Kim, H.J.; Cho, H.; Alexander, R.; Patterson, H.C.; Gu, M.; Lo, K.A.; Xu, D.; Goh, V.J.; Nguyen, L.N.; Chai, X.; et al. MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes 2014, 63, 4045–4056. [Google Scholar] [CrossRef]
- Fu, T.; Seok, S.; Choi, S.; Huang, Z.; Suino-Powell, K.; Xu, H.E.; Kemper, B.; Kemper, J.K. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell Biol. 2014, 34, 4130–4142. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Siegel, F.; Kipschull, S.; Haas, B.; Frohlich, H.; Meister, G.; Pfeifer, A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 2013, 4, 1769. [Google Scholar] [CrossRef]
- Davis, C.D.; Ross, S.A. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr. Rev. 2008, 66, 477–482. [Google Scholar] [CrossRef]
- Mori, M.A.; Thomou, T.; Boucher, J.; Lee, K.Y.; Lallukka, S.; Kim, J.K.; Torriani, M.; Yki-Järvinen, H.; Grinspoon, S.K.; Cypess, A.M.; et al. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J. Clin. Investig. 2014, 124, 3339–3351. [Google Scholar] [CrossRef] [PubMed]
- Price, N.L.; Fernandez-Hernando, C. miRNA regulation of white and brown adipose tissue differentiation and function. Biochim. Biophys. Acta 2016, 1861, 2104–2110. [Google Scholar] [CrossRef]
- Zhang, J.; Kay, M.K.; Park, M.H.; Meruvu, S.; Powell, C.; Choudhury, M. LncRNA DLEU2 regulates sirtuins and mitochondrial respiratory chain complex IV: A novel pathway in obesity and offspring’s health. Int. J. Obes. 2022, 46, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Ziqubu, K.; Dludla, P.V.; Mthembu, S.X.H.; Nkambule, B.B.; Mabhida, S.E.; Jack, B.U.; Nyambuya, T.M.; Mazibuko-Mbeje, S.E. An insight into brown/beige adipose tissue whitening, a metabolic complication of obesity with the multifactorial origin. Front. Endocrinol. 2023, 14, 1114767. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, M.; Tong, M.; Yang, L.; Pang, L.; Chen, L.; Xu, G.; Chi, X.; Hong, Q.; Ni, Y.; et al. miR-148a is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Sci. Rep. 2015, 5, 9930. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Wilfred, B.R.; Wang, W.X.; Nelson, P.T. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007, 91, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167–107175. [Google Scholar] [CrossRef]
- Carpentier, A.C.; Blondin, D.P.; Virtanen, K.A.; Richard, D.; Haman, F.; Turcotte, E.E. Brown Adipose Tissue Energy Metabolism in Humans. Front. Endocrinol. 2018, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Xiao, W.; You, L.; Zhang, F.; Cao, X.; Feng, J.; Shen, D.; Li, Y.; Wang, Y.; Ji, C.; et al. Age-induced oxidative stress impairs adipogenesis and thermogenesis in brown fat. FEBS J. 2019, 286, 2753–2768. [Google Scholar] [CrossRef]
- Ohtomo, T.; Ino, K.; Miyashita, R.; Chigira, M.; Nakamura, M.; Someya, K.; Inaba, N.; Fujita, M.; Takagi, M.; Yamada, J. Chronic high-fat feeding impairs adaptive induction of mitochondrial fatty acid combustion-associated proteins in brown adipose tissue of mice. Biochem. Biophys. Rep. 2017, 10, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Kenmochi, H.; Miyashita, Y.; Sasaki, M.; Ojima, M.; Sasahara, M.; Koya, D.; Tsuneki, H.; Sasaoka, T. Spironolactone improves glucose and lipid metabolism by ameliorating hepatic steatosis and inflammation and suppressing enhanced gluconeogenesis induced by high-fat and high-fructose diet. Endocrinology 2010, 151, 2040–2049. [Google Scholar] [CrossRef]
- Zhang, J.; Powell, C.A.; Kay, M.K.; Sonkar, R.; Meruvu, S.; Choudhury, M. Effect of Chronic Western Diets on Non-Alcoholic Fatty Liver of Male Mice Modifying the PPAR-γ Pathway via miR-27b-5p Regulation. Int. J. Mol. Sci. 2021, 22, 1822. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Tabak, L.A. Policy: NIH plans to enhance reproducibility. Nature 2014, 505, 612–613. [Google Scholar] [CrossRef]
- Kotzbeck, P.; Giordano, A.; Mondini, E.; Murano, I.; Severi, I.; Venema, W.; Cecchini, M.P.; Kershaw, E.E.; Barbatelli, G.; Haemmerle, G.; et al. Brown adipose tissue whitening leads to brown adipocyte death and adipose tissue inflammation. J. Lipid Res. 2018, 59, 784–794. [Google Scholar] [CrossRef]
- Leitner, B.P.; Huang, S.; Brychta, R.J.; Duckworth, C.J.; Baskin, A.S.; McGehee, S.; Tal, I.; Dieckmann, W.; Gupta, G.; Kolodny, G.M.; et al. Mapping of human brown adipose tissue in lean and obese young men. Proc. Natl. Acad. Sci. USA 2017, 114, 8649–8654. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, B.R.; McLennan, S.V.; Yee, C.; Twigg, S.M.; Williams, P.F. The Effect of a Sustained High-Fat Diet on the Metabolism of White and Brown Adipose Tissue and Its Impact on Insulin Resistance: A Selected Time Point Cross-Sectional Study. Int. J. Mol. Sci. 2021, 22, 13639. [Google Scholar] [CrossRef] [PubMed]
- Dickson, L.M.; Gandhi, S.; Layden, B.T.; Cohen, R.N.; Wicksteed, B. Protein kinase A induces UCP1 expression in specific adipose depots to increase energy expenditure and improve metabolic health. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R79–R88. [Google Scholar] [CrossRef]
- Moreno-Fernandez, S.; Garces-Rimon, M.; Uranga, J.A.; Astier, J.; Landrier, J.F.; Miguel, M. Expression enhancement in brown adipose tissue of genes related to thermogenesis and mitochondrial dynamics after administration of pepsin egg white hydrolysate. Food Funct. 2018, 9, 6599–6607. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gu, H.; Gan, L.; Xu, Y.; Feng, F.; Saeed, M.; Sun, C. Reducing Smad3/ATF4 was essential for Sirt1 inhibiting ER stress-induced apoptosis in mice brown adipose tissue. Oncotarget 2017, 8, 9267–9279. [Google Scholar] [CrossRef]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shree, N.; Meruvu, S.; Park, M.H.; Choudhury, M. High-Fat-High-Fructose Diet Elicits Brown Adipocyte Dysfunction through miRNA-103 Induced miRNA Biogenesis Pathway. Obesities 2024, 4, 93-105. https://doi.org/10.3390/obesities4020010
Shree N, Meruvu S, Park MH, Choudhury M. High-Fat-High-Fructose Diet Elicits Brown Adipocyte Dysfunction through miRNA-103 Induced miRNA Biogenesis Pathway. Obesities. 2024; 4(2):93-105. https://doi.org/10.3390/obesities4020010
Chicago/Turabian StyleShree, Nitya, Sunitha Meruvu, Min Hi Park, and Mahua Choudhury. 2024. "High-Fat-High-Fructose Diet Elicits Brown Adipocyte Dysfunction through miRNA-103 Induced miRNA Biogenesis Pathway" Obesities 4, no. 2: 93-105. https://doi.org/10.3390/obesities4020010
APA StyleShree, N., Meruvu, S., Park, M. H., & Choudhury, M. (2024). High-Fat-High-Fructose Diet Elicits Brown Adipocyte Dysfunction through miRNA-103 Induced miRNA Biogenesis Pathway. Obesities, 4(2), 93-105. https://doi.org/10.3390/obesities4020010





