Gut Microbiota Profiles of Children with Obesity or Metabolic Syndrome: Body Mass Index Is a Lead Actor
Abstract
:1. Introduction
2. Methods
2.1. Ethics Statement
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Dietary Intake and Data Analysis
2.5. Microbiome Analysis
2.6. Statistical Analysis
3. Results
3.1. Baseline Characteristics of Participants
3.2. Analysis of Gut Microbiota Composition
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hales, C.M.; Carroll, M.D.; Fryar, C.D.; Ogden, C.L. Prevalence of Obesity Among Adults and Youth: United States, 2015–2016. NCHS Data Brief 2017, 288, 1–8. [Google Scholar]
- Zawada, A.; Rychter, A.M.; Ratajczak, A.E.; Lisiecka-Masian, A.; Dobrowolska, A.; Krela-Kazmierczak, I. Does Gut-Microbiome Interaction Protect against Obesity and Obesity-Associated Metabolic Disorders? Microorganisms 2020, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.B.; Smith, M.S. Obesity Statistics. Prim. Care 2016, 43, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Janssen, F.; Bardoutsos, A.; Vidra, N. Obesity Prevalence in the Long-Term Future in 18 European Countries and in the USA. Obes. Facts 2020, 13, 514–527. [Google Scholar] [CrossRef]
- Magge, S.N.; Goodman, E.; Armstrong, S.C.; Daniels, S.; Corkins, M.; de Ferranti, S.; Golden, N.H.; Kim, J.H.; Schwarzenberg, S.J.; Sills, I.N.; et al. The Metabolic Syndrome in Children and Adolescents: Shifting the Focus to Cardiometabolic Risk Factor Clustering. Pediatrics 2017, 140, e20171603. [Google Scholar] [CrossRef]
- Farpour-Lambert, N.J.; Baker, J.L.; Hassapidou, M.; Holm, J.C.; Nowicka, P.; O’Malley, G.; Weiss, R. Childhood Obesity Is a Chronic Disease Demanding Specific Health Care—A Position Statement from the Childhood Obesity Task Force (COTF) of the European Association for the Study of Obesity (EASO). Obes. Facts 2015, 8, 342–349. [Google Scholar] [CrossRef]
- Carrizales-Sanchez, A.K.; Garcia-Cayuela, T.; Hernandez-Brenes, C.; Senes-Guerrero, C. Gut microbiota associations with metabolic syndrome and relevance of its study in pediatric subjects. Gut Microbes 2021, 13, 1960135. [Google Scholar] [CrossRef]
- Weihe, P.; Weihrauch-Bluher, S. Metabolic Syndrome in Children and Adolescents: Diagnostic Criteria, Therapeutic Options and Perspectives. Curr. Obes. Rep. 2019, 8, 472–479. [Google Scholar] [CrossRef]
- Almeida, A.; Mitchell, A.L.; Boland, M.; Forster, S.C.; Gloor, G.B.; Tarkowska, A.; Lawley, T.D.; Finn, R.D. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504. [Google Scholar] [CrossRef]
- Davis, C.D. The Gut Microbiome and Its Role in Obesity. Nutr. Today 2016, 51, 167–174. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Martinez, J.A. Diet, Gut Microbiota, and Obesity: Links with Host Genetics and Epigenetics and Potential Applications. Adv. Nutr. 2019, 10 (Suppl. S1), S17–S30. [Google Scholar] [CrossRef] [PubMed]
- King, C.H.; Desai, H.; Sylvetsky, A.C.; LoTempio, J.; Ayanyan, S.; Carrie, J.; Crandall, K.A.; Fochtman, B.C.; Gasparyan, L.; Gulzar, N.; et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS ONE 2019, 14, e0206484. [Google Scholar] [CrossRef] [PubMed]
- Dabke, K.; Hendrick, G.; Devkota, S. The gut microbiome and metabolic syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef] [PubMed]
- Marotz, C.A.; Zarrinpar, A. Treating Obesity and Metabolic Syndrome with Fecal Microbiota Transplantation. Yale J. Biol. Med. 2016, 89, 383–388. [Google Scholar]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. A review on gut microbiota: A central factor in the pathophysiology of obesity. Lipids Health Dis. 2021, 20, 65. [Google Scholar] [CrossRef]
- Abenavoli, L.; Scarpellini, E.; Colica, C.; Boccuto, L.; Salehi, B.; Sharifi-Rad, J.; Aiello, V.; Romano, B.; De Lorenzo, A.; Izzo, A.A.; et al. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients 2019, 11, 2690. [Google Scholar] [CrossRef]
- Mazidi, M.; Rezaie, P.; Kengne, A.P.; Mobarhan, M.G.; Ferns, G.A. Gut microbiome and metabolic syndrome. Diabetes Metab. Syndr. 2016, 10 (Suppl. S1), S150–S157. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Boer, C.G.; Beth, S.A.; van der Wal, P.; Kiefte-De Jong, J.C.; Jansen, M.A.E.; Konstantinov, S.R.; Peppelenbosch, M.P.; Hays, J.P.; Jaddoe, V.W.V.; et al. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci. Rep. 2020, 10, 1040. [Google Scholar] [CrossRef]
- Ahrens, W.; Moreno, L.A.; Marild, S.; Molnar, D.; Siani, A.; De Henauw, S.; Bohmann, J.; Gunther, K.; Hadjigeorgiou, C.; Iacoviello, L.; et al. Metabolic syndrome in young children: Definitions and results of the IDEFICS study. Int. J. Obes. 2014, 38 (Suppl. S2), S4–S14. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; Qiu, Y.; Cao, Y.; Zhang, S.; Lu, L.; Kofonow, J.M.; Robertson, C.E.; Liu, Y.; Wang, H.; Levens, C.L.; et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene 2022, 41, 1269–1280. [Google Scholar] [CrossRef]
- Vickery, T.W.; Armstrong, M.; Kofonow, J.M.; Robertson, C.E.; Kroehl, M.E.; Reisdorph, N.A.; Ramakrishnan, V.R.; Frank, D.N. Altered tissue specialized pro-resolving mediators in chronic rhinosinusitis. Prostaglandins Leukot. Essent. Fat. Acids 2021, 164, 102218. [Google Scholar] [CrossRef]
- Soderborg, T.K.; Clark, S.E.; Mulligan, C.E.; Janssen, R.C.; Babcock, L.; Ir, D.; Young, B.; Krebs, N.; Lemas, D.J.; Johnson, L.K.; et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat. Commun. 2018, 9, 4462. [Google Scholar] [CrossRef] [PubMed]
- Homo Sapiens UCSC Hg19 Human Genome Sequence from iGenome. Available online: https://support.illumina.com/sequencing/sequencing_software/igenome.html (accessed on 8 December 2014).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glockner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Robertson, C.E.; Harris, J.K.; Wagner, B.D.; Granger, D.; Browne, K.; Tatem, B.; Feazel, L.M.; Park, K.; Pace, N.R.; Frank, D.N. Explicet: Graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics 2013, 29, 3100–3101. [Google Scholar] [CrossRef] [PubMed]
- A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019.
- Anderson, M.J.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Oksanen, J.B.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.; et al. Vegan: Community Ecology Package. R Package version 2.5-4. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 16 July 2023).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Grummer-Strawn, L.M.; Reinold, C.; Krebs, N.F.; Centers for Disease Control and Prevention. Use of World Health Organization and CDC growth charts for children aged 0–59 months in the United States. MMWR Recomm. Rep. 2010, 59, 1–15. [Google Scholar] [PubMed]
- de Onis, M.; Onyango, A.W.; Borghi, E.; Siyam, A.; Nishida, C.; Siekmann, J. Development of a WHO growth reference for school-aged children and adolescents. Bull. World Health Organ. 2007, 85, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.J.; Bellizzi, M.C.; Flegal, K.M.; Dietz, W.H. Establishing a standard definition for child overweight and obesity worldwide: International survey. BMJ Clin. Res. Ed 2000, 320, 1240–1243. [Google Scholar] [CrossRef]
- Joseph, N.; Clayton, J.B.; Hoops, S.L.; Linhardt, C.A.; Mohd Hashim, A.; Mohd Yusof, B.N.; Kumar, S.; Amin Nordin, S. Alteration of the Gut Microbiome in Normal and Overweight School Children from Selangor with Lactobacillus Fermented Milk Administration. Evol. Bioinform. Online 2020, 16, 1176934320965943. [Google Scholar] [CrossRef]
- Wutthi-In, M.; Cheevadhanarak, S.; Yasom, S.; Kerdphoo, S.; Thiennimitr, P.; Phrommintikul, A.; Chattipakorn, N.; Kittichotirat, W.; Chattipakorn, S. Gut Microbiota Profiles of Treated Metabolic Syndrome Patients and their Relationship with Metabolic Health. Sci. Rep. 2020, 10, 10085. [Google Scholar] [CrossRef]
- Qin, Q.; Yan, S.; Yang, Y.; Chen, J.; Li, T.; Gao, X.; Yan, H.; Wang, Y.; Wang, J.; Wang, S.; et al. A Metagenome-Wide Association Study of the Gut Microbiome and Metabolic Syndrome. Front. Microbiol. 2021, 12, 682721. [Google Scholar] [CrossRef] [PubMed]
- Haro, C.; Garcia-Carpintero, S.; Alcala-Diaz, J.F.; Gomez-Delgado, F.; Delgado-Lista, J.; Perez-Martinez, P.; Rangel Zuniga, O.A.; Quintana-Navarro, G.M.; Landa, B.B.; Clemente, J.C.; et al. The gut microbial community in metabolic syndrome patients is modified by diet. J. Nutr. Biochem. 2016, 27, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Penders, J.; Shi, Z.; Ren, H.; Cai, K.; Fang, C.; Ding, Q.; Thijs, C.; Blaak, E.E.; Stehouwer, C.D.A.; et al. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome 2019, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Nirmalkar, K.; Murugesan, S.; Pizano-Zarate, M.L.; Villalobos-Flores, L.E.; Garcia-Gonzalez, C.; Morales-Hernandez, R.M.; Nunez-Hernandez, J.A.; Hernandez-Quiroz, F.; Romero-Figueroa, M.D.S.; Hernandez-Guerrero, C.; et al. Gut Microbiota and Endothelial Dysfunction Markers in Obese Mexican Children and Adolescents. Nutrients 2018, 10, 2009. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Becerra, L.; Cornejo-Granados, F.; Garcia-Lopez, R.; Valdez-Lara, A.; Bikel, S.; Canizales-Quinteros, S.; Lopez-Contreras, B.E.; Mendoza-Vargas, A.; Nielsen, H.; Ochoa-Leyva, A. Metatranscriptomic analysis to define the Secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children. Microb. Cell Fact. 2020, 19, 61. [Google Scholar] [CrossRef]
- Parekh, P.J.; Balart, L.A.; Johnson, D.A. The Influence of the Gut Microbiome on Obesity, Metabolic Syndrome and Gastrointestinal Disease. Clin. Transl. Gastroenterol. 2015, 6, e91. [Google Scholar] [CrossRef]
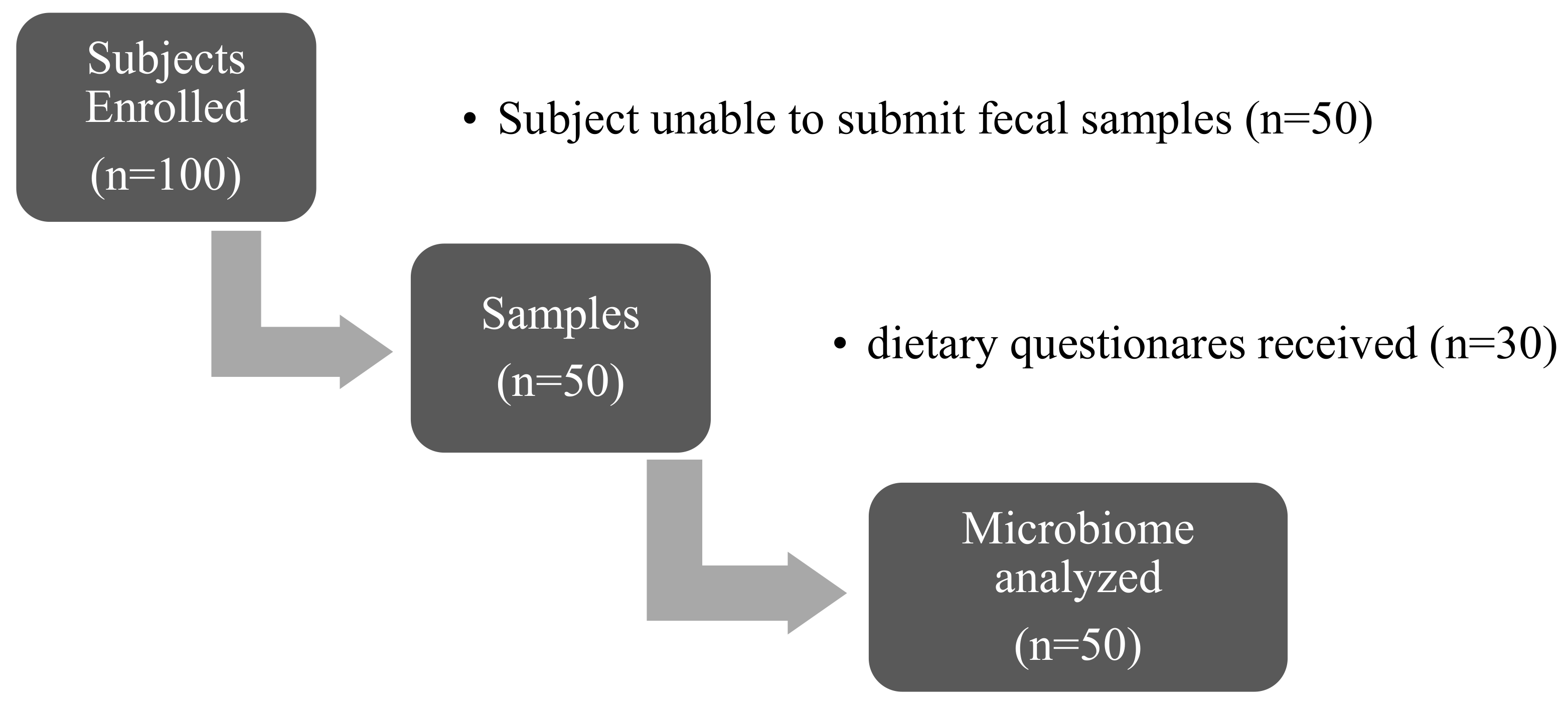
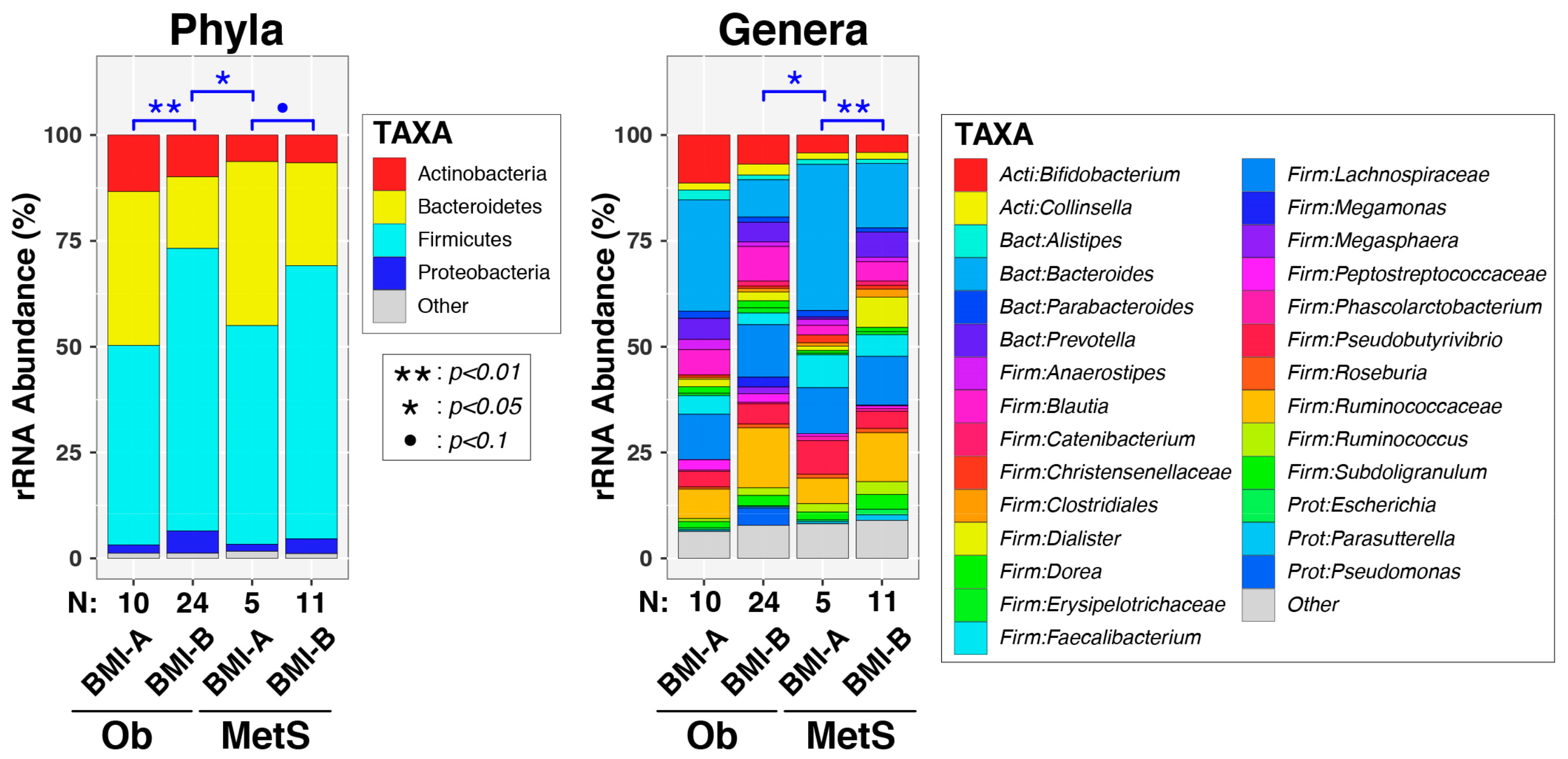
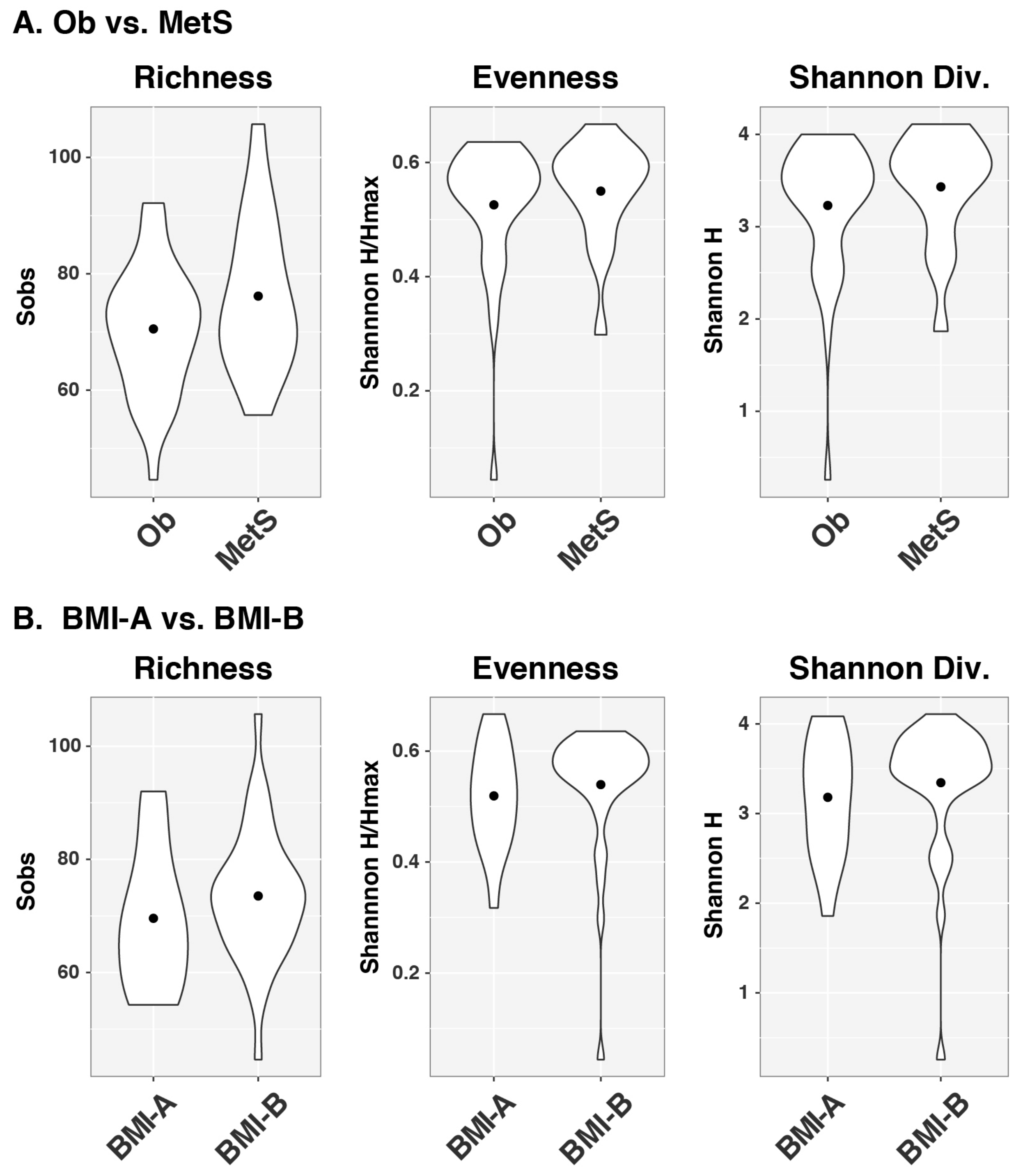
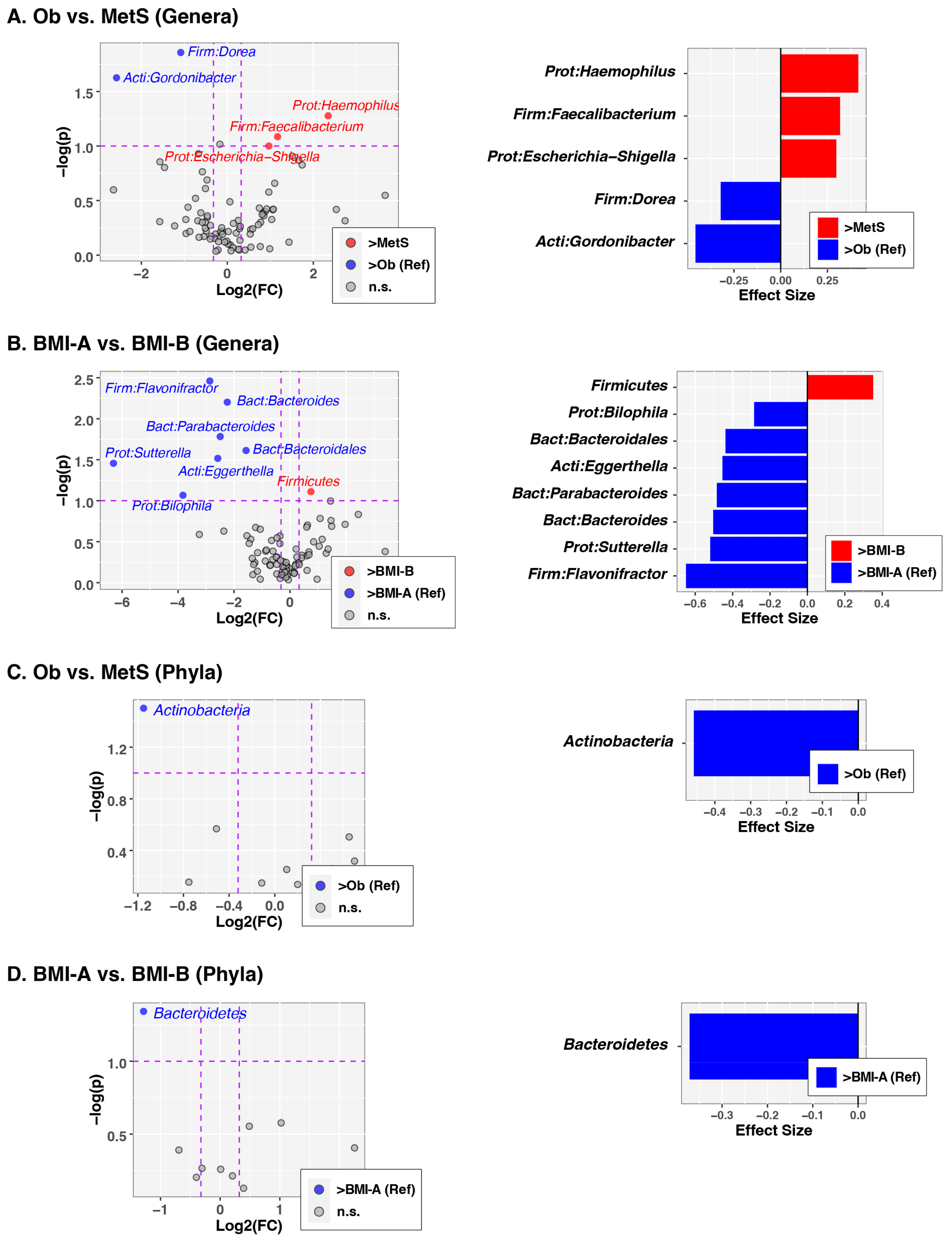
| Variable | OB (N = 33) | MetS (N = 17) | p Value |
|---|---|---|---|
| Age (Years) | 14.7 ± 2.3 | 14.6 ± 2.5 | 0.9090t |
| Males | 18 (54.6%) | 11 (64.7%) | 0.490f |
| White Latino Other | 20 (60.6%) 7 (21.2%) 6 (18.2%) | 6 (35.3%) 6 (35.3%) 5 (29.41%) | 0.237f |
| Body mass index (kg/m2) | 32.5 ± 4.3 | 35.3 ± 7.8 | 0.1060t |
| Waist circumference (cm) | 102.7 ± 15.5 | 105.5 ± 24.6 | 0.83t |
| Height (cm) | 164.4 ± 8.7 | 162.6 ± 11.6 | 0.5477t |
| Triglycerides | 115.3 ± 47.6 | 218.2 ± 127.5 | 0.0007t |
| HDL-cholesterol | 47.0 ± 7.4 | 38.8 ± 13.6 | 0.0158t |
| Systolic BPi | 0.91 ± 0.06 | 1.0 ± 0.1 | 0.0008t |
| Diastolic BPi | 0.84 ± 0.09 | 0.94 ± 0.12 | 0.0021t |
| Hemoglobin A1C (%) | 5.5 ± 0.3 | 5.8 ± 0.5 | 0.0229t |
| Family history of diabetes mellitus | 9 (27.3%) | 10 (58.8%) | 0.029f |
| Variable | Ob (N = 20) | MetS (N = 10) | t |
|---|---|---|---|
| Age (years) | 14 ± 2.3 | 16.2 ± 1.4 | 0.01 |
| Male | 11(55%) | 5 (50%) | 0.99f |
| Latino | 7 (35%) | 2 (20%) | 0.55f |
| BMI | 31.9 (IQR 28.7, 34) | 33.7 (IQR 32.6, 35.8) | 0.12k |
| Average Intake Per Day | |||
| Kilocalories | 1670.9 ± 555.6 | 1557.4 ± 380.7 | 0.57t |
| Protein (g) | 68.2 ± 18.6 | 69.6 ± 12.8 | 0.83t |
| Carbohydrate (g) | 210.1 (IQR 133.7, 289.4) | 190.9 (IQR 166.8, 199.2) | 0.96k |
| Sugar, Total (g) | 88.0 (IQR 31.5, 109.4) | 68.4 (IQR 46.1, 76.4) | 0.92k |
| Total Dietary Fiber (g) | 12.5 ± 4.8 | 15.7 ± 6.6 | 0.14t |
| Fat (g) | 62.7 (IQR 47.2, 78.8) | 55.9 (IQR 51.6, 55.6) | 0.21k |
| Saturated fat (g) | 23.1 ± 9.1 | 18.6 ± 4.2 | 0.16t |
| Sodium (mg) | 2656.9 ± 765.7 | 2563.4 ± 471.4 | 0.73t |
| Potassium (mg) | 1383.7 ± 458.9 | 1766.5 ± 520.4 | 0.049t |
| Vitamin D (IU) | 100.5 (IQR 39.6, 163.7) | 170.5 (IQR 146.3, 174.2) | 0.02k |
| Folate (ug) | 216.7 (IQR 122.5, 305.4) | 409.6 (IQR 273.5, 476.5) | 0.005k |
| Caffeine (mg) | 15.7 (IQR 0, 22.1) | 1.2 (IQR 0, 3.6) | 0.19k |
| PERMANOVA p-Value | |||
|---|---|---|---|
| Variable | Genus | Phylum | Notes |
| Disease Category | 0.32 | 0.77 | Ob vs. MetS |
| BMI | 0.013 | 0.0028 | BMI < 30 vs. BMI ≥ 30 |
| Age | 0.054 | 0.31 | Years |
| Sex | 0.75 | 0.93 | F vs. M |
| Ethnicity | 0.046 | 0.052 | Asian, Black, Latino, White, and other |
| SBPI | 0.55 | 0.56 | High vs. Low SBPI (SBPI > 1) |
| DBPI | 0.36 | 0.59 | High vs. Low DBPI (DBPI > 1) |
| abBP | 0.24 | 0.68 | Low BP vs. High (SBPI OR DBPI) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gathungu, G.N.; Frank, D.N.; Chawla, A.; Robertson, C.E.; LaComb, J.F.; Markarian, K.; Burghard, B.M.; Woroniecki, R. Gut Microbiota Profiles of Children with Obesity or Metabolic Syndrome: Body Mass Index Is a Lead Actor. Obesities 2023, 3, 253-264. https://doi.org/10.3390/obesities3030021
Gathungu GN, Frank DN, Chawla A, Robertson CE, LaComb JF, Markarian K, Burghard BM, Woroniecki R. Gut Microbiota Profiles of Children with Obesity or Metabolic Syndrome: Body Mass Index Is a Lead Actor. Obesities. 2023; 3(3):253-264. https://doi.org/10.3390/obesities3030021
Chicago/Turabian StyleGathungu, Grace N., Daniel N. Frank, Anupama Chawla, Charles E. Robertson, Joseph F. LaComb, Katherine Markarian, Brianna M. Burghard, and Robert Woroniecki. 2023. "Gut Microbiota Profiles of Children with Obesity or Metabolic Syndrome: Body Mass Index Is a Lead Actor" Obesities 3, no. 3: 253-264. https://doi.org/10.3390/obesities3030021
APA StyleGathungu, G. N., Frank, D. N., Chawla, A., Robertson, C. E., LaComb, J. F., Markarian, K., Burghard, B. M., & Woroniecki, R. (2023). Gut Microbiota Profiles of Children with Obesity or Metabolic Syndrome: Body Mass Index Is a Lead Actor. Obesities, 3(3), 253-264. https://doi.org/10.3390/obesities3030021







