Effects of High-Fat Diet on the Gut Microbiota of Renalase Gene Knockout Mice
Abstract
1. Introduction
2. Materials and Methods
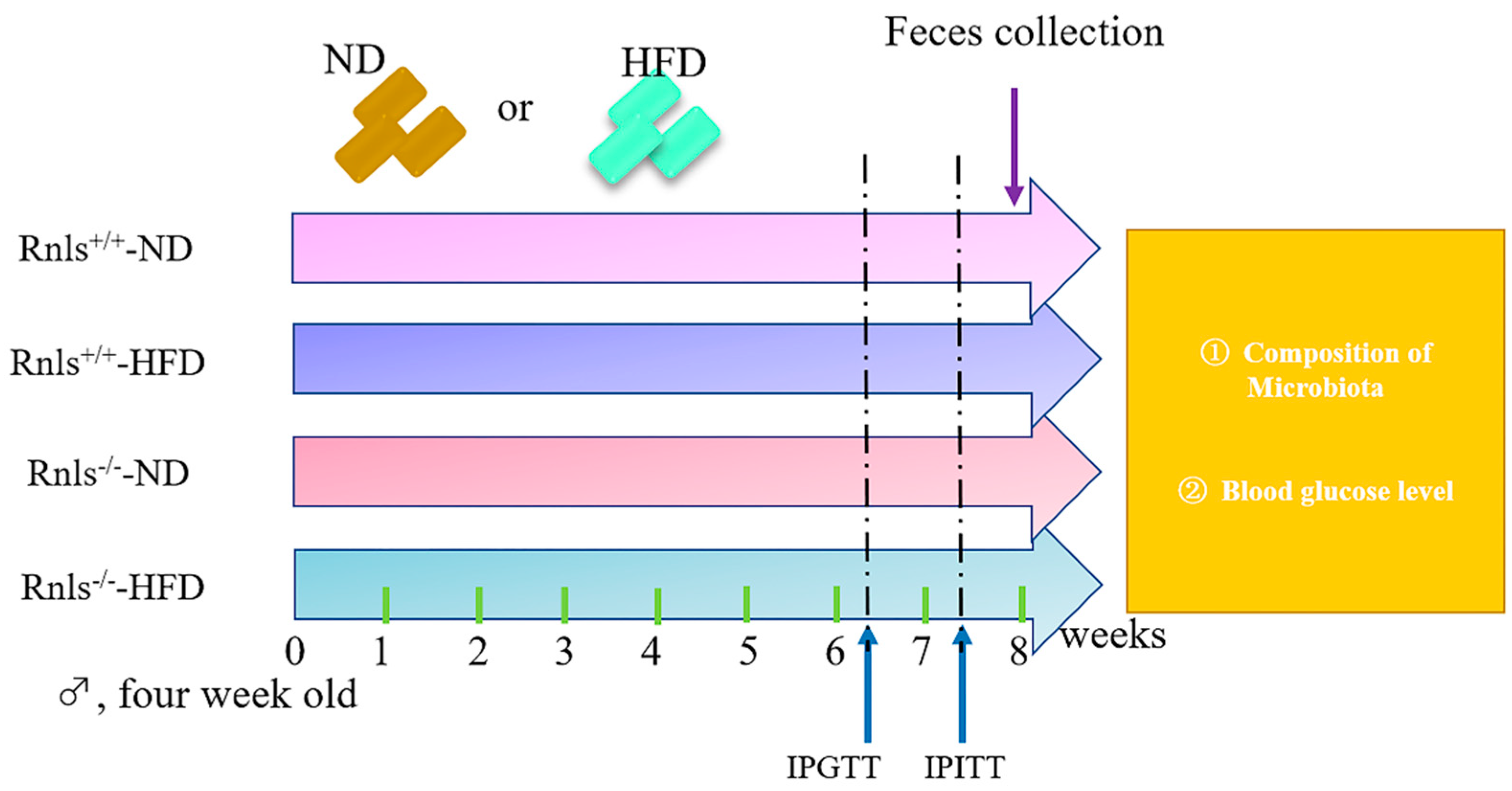
2.1. Mice Management and Experimental Design
2.2. Body Weight
2.3. IPGTT and IPITT
2.4. Feces Collection
2.5. Feces DNA Extraction and 16S rRNA High Throughput Sequencing
2.6. Mice Euthanasia
2.7. Statistical Analysis
3. Results
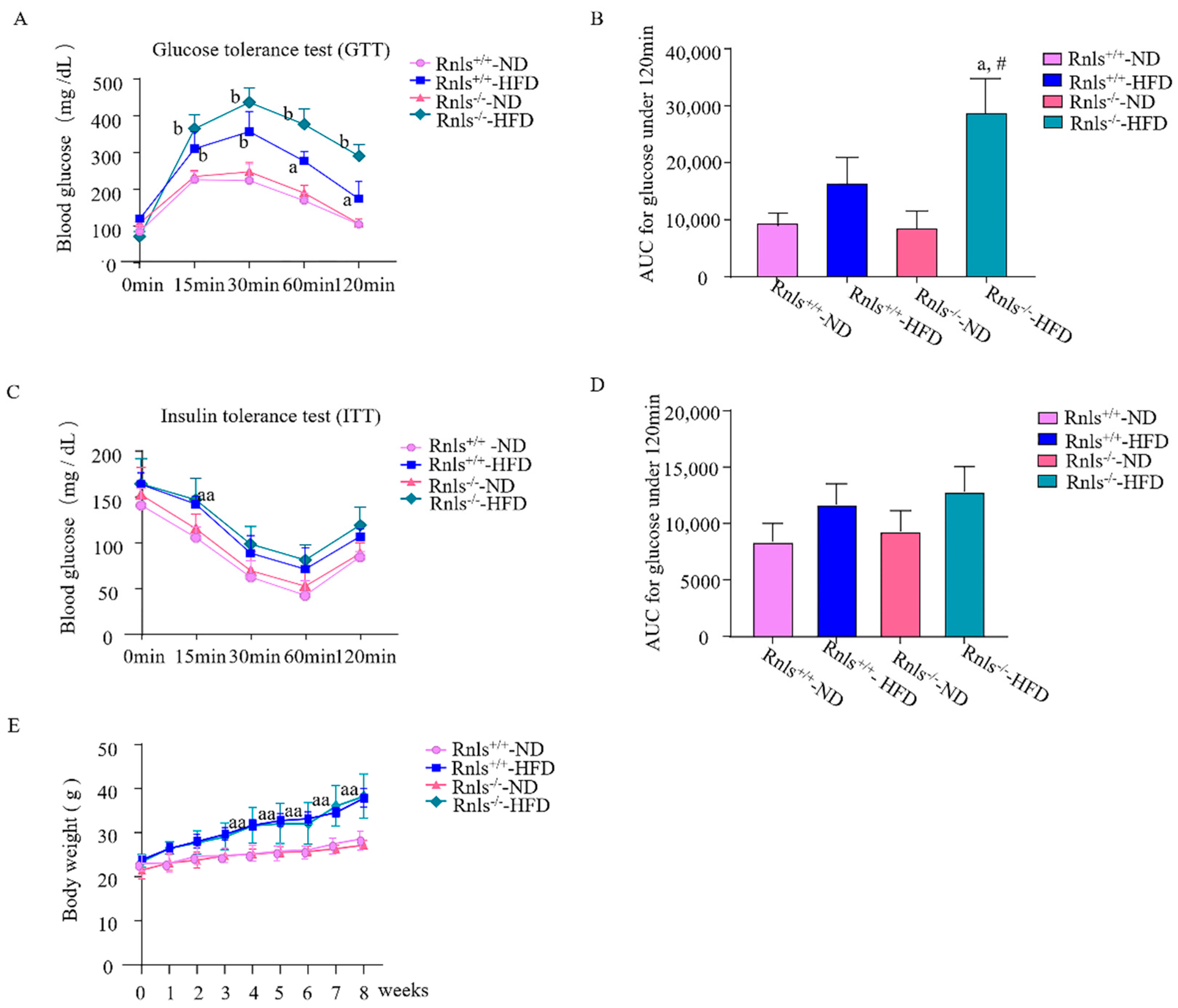
3.1. Rnls−/− Mice Fed with HFD for 8 Weeks Exhibited Impaired Glucose Tolerance
3.2. Individual Taxa Richness and Evenness of Microbiota in Rnls+/+ and Rnls−/− Mice
3.3. Microbial Distribution in Rnls+/+ and Rnls−/− Mice
3.4. Overall Compositions of Microbiota in Rnls+/+ and Rnls−/− Mice
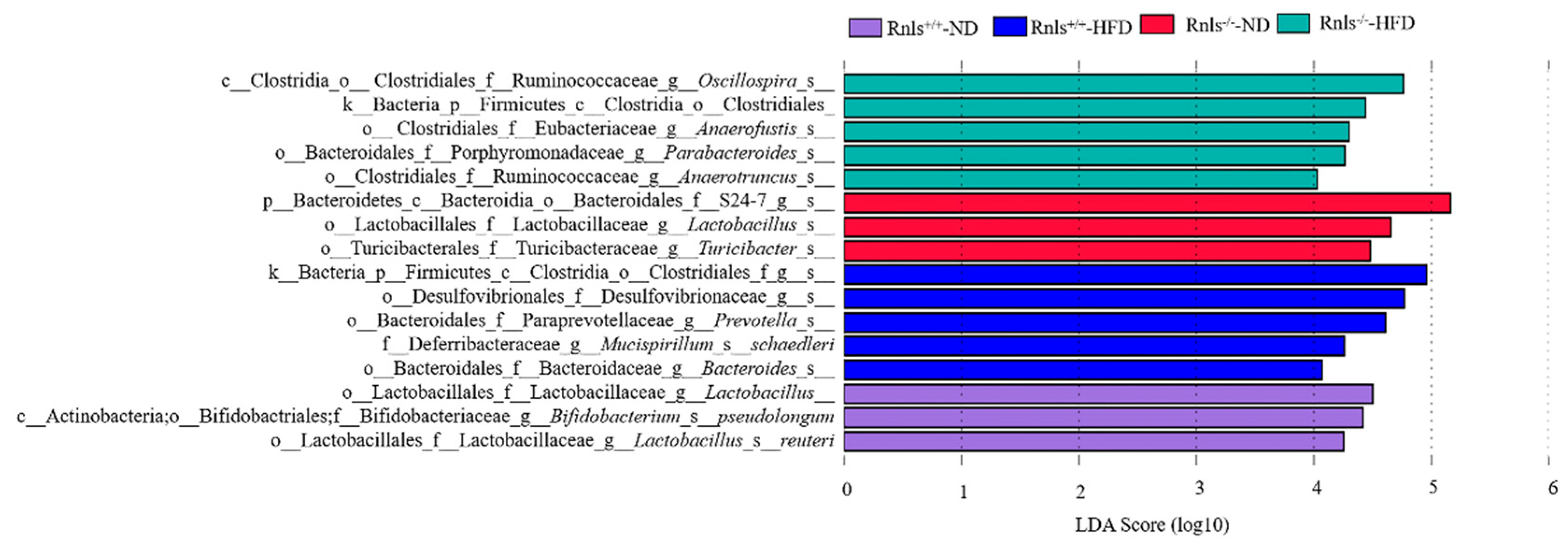
3.5. Significant Differences in Species Were Observed in the Microbiota of Rnls+/+ and Rnls−/− Mice
3.6. Dominant Patterns of Microbiota Composition in Rnls+/+ and Rnls−/− Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Xu, J.; Li, G.; Wang, P.; Velazquez, H.; Yao, X.; Li, Y.; Wu, Y.; Peixoto, A.; Crowley, S.; Desir, G.V. Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J. Clin. Investig. 2005, 115, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, L.; Wang, X.; Wang, Y.; Zhang, Q.; Liu, W. Renalase contributes to protection against renal fibrosis via inhibiting oxidative stress in rats. Int. Urol. Nephrol. 2018, 50, 1347–1354. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, Z.; Lin, M.; Huang, R.; Liang, Z.; Huang, W.; Jiang, W. Renalase Protects the Cardiomyocytes of Sprague-Dawley Rats Against Ischemia and Reperfusion Injury by Reducing Myocardial Cell Necrosis and Apoptosis. Kidney Blood Press. Res. 2015, 40, 215–222. [Google Scholar] [CrossRef]
- Pointer, T.; Gorelick, F.; Desir, G. Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease. Cells 2021, 10, 2006. [Google Scholar] [CrossRef]
- Tokinoya, K.; Sekine, N.; Aoki, K.; Ono, S.; Kuji, T.; Sugasawa, T.; Yoshida, Y.; Takekoshi, K. Effects of renalase deficiency on liver fibrosis markers in a nonalcoholic steatohepatitis mouse model. Mol. Med. Rep. 2021, 23, 210. [Google Scholar] [CrossRef]
- Aoki, K.; Yanazawa, K.; Tokinoya, K.; Sugasawa, T.; Suzuki, T.; Yoshida, Y.; Nakano, T.; Omi, N.; Kawakami, Y.; Takekoshi, K. Renalase is localized to the small intestine crypt and expressed upon the activation of NF-κB p65 in mice model of fasting-induced oxidative stress. Life Sci. 2020, 267, 118904. [Google Scholar] [CrossRef]
- Luc, K.; Schramm-Luc, A.; Guzik, T.J.; Mikolajczyk, T.P. Oxidative stress and inflammatory markers in prediabetes and dia-betes. J. Physiol. Pharmacol. 2019, 70, 70. [Google Scholar] [CrossRef]
- Vassalle, C.; Gaggini, M. Type 2 Diabetes and Oxidative Stress and Inflammation: Pathophysiological Mechanisms and Possible Therapeutic Options. Antioxidants 2022, 11, 953. [Google Scholar] [CrossRef]
- Lafferty, R.A.; O’Harte, F.P.M.; Irwin, N.; Gault, V.A.; Flatt, P.R. Proglucagon-Derived Peptides as Therapeutics. Front. Endocrinol. 2021, 12, 689678. [Google Scholar] [CrossRef]
- Teimoori, B.; Moradi-Shahrebabak, M.; Rezaei, M.; Mohammadpour-Gharehbagh, A.; Salimi, S. Renalase rs10887800 polymorphism is associated with severe pre-eclampsia in southeast Iranian women. J. Cell. Biochem. 2018, 120, 3277–3285. [Google Scholar] [CrossRef]
- Buraczynska, M.; Gwiazda-Tyndel, K.; Drop, B.; Zaluska, W. Renalase gene Glu37Asp polymorphism affects susceptibility to diabetic retinopathy in type 2 diabetes mellitus. Geol. Rundsch. 2021, 58, 1595–1602. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, W.; Wu, Y.; Li, X.; Zhang, S.; Feng, Y.; Lu, R.; Sun, L. Association of renalase gene polymorphisms with the risk of hypertensive disorders of pregnancy in northeastern Han Chinese population. Gynecol. Endocrinol. 2020, 36, 986–990. [Google Scholar] [CrossRef]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef]
- Thingholm, L.B.; Rühlemann, M.C.; Koch, M.; Fuqua, B.; Laucke, G.; Boehm, R.; Bang, C.; Franzosa, E.A.; Hübenthal, M.; Rahnavard, G.; et al. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe 2019, 26, 252–264.e10. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Yang, F.; Zhu, W.-J.; Edirisuriya, P.; Ai, Q.; Nie, K.; Ji, X.-M.; Li, Y.; Zhou, K. Beneficial effects of a combination of Clostridium cochlearium and Lactobacillus acidophilus on body weight gain, insulin sensitivity, and gut microbiota in high-fat diet–induced obese mice. Nutrition 2021, 93, 111439. [Google Scholar] [CrossRef]
- Yoon, H.S.; Cho, C.H.; Yun, M.S.; Jang, S.J.; You, H.J.; Kim, J.-H.; Han, D.; Cha, K.H.; Moon, S.H.; Lee, K.; et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat. Microbiol. 2021, 6, 563–573. [Google Scholar] [CrossRef]
- Davenport, E.R. Elucidating the role of the host genome in shaping microbiome composition. Gut Microbes 2016, 7, 178–184. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Clark, A.G.; Ley, R.E. The Relationship Between the Human Genome and Microbiome Comes into View. Annu. Rev. Genet. 2017, 51, 413–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tong, X.; Zhu, J.; Tian, L.; Jie, Z.; Zou, Y.; Lin, X.; Liang, H.; Li, W.; Ju, Y.; et al. Metagenome-genome-wide association studies reveal human genetic impact on the oral microbiome. Cell Discov. 2021, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, S.; Zhong, H.; Tong, X.; Jie, Z.; Ding, Q.; Wang, D.; Guo, R.; Xiao, L.; Xu, X.; et al. A genome-wide association study for gut metagenome in Chinese adults illuminates complex diseases. Cell Discov. 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Zajac, D.J.; Green, S.J.; Johnson, L.A.; Estus, S. APOE genetics influence murine gut microbiome. Sci. Rep. 2022, 12, 1906. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Rani, K.; Datt, C. Molecular link between dietary fibre, gut microbiota and health. Mol. Biol. Rep. 2020, 47, 6229–6237. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef]
- Nagy, C.; Einwallner, E. Study of In Vivo Glucose Metabolism in High-fat Diet-fed Mice Using Oral Glucose Tolerance Test (OGTT) and Insulin Tolerance Test (ITT). J. Vis. Exp. 2018, 131, e56672. [Google Scholar] [CrossRef]
- Beck, D.; Foster, J. Machine Learning Techniques Accurately Classify Microbial Communities by Bacterial Vaginosis Characteristics. PLoS ONE 2014, 9, e87830. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Kaul, A.; Davidov, O.; Peddada, S.D. Structural zeros in high-dimensional data with applications to microbiome studies. Biostatistics 2017, 18, 422–433. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, D.; Yu, X.; Ye, A.; Xu, C.; Li, X.; Geng, W.; Zhu, L. Profiling of gut microbial dysbiosis in adults with myeloid leukemia. FEBS Open Bio 2021, 11, 2050–2059. [Google Scholar] [CrossRef]
- Czubilińska-Łada, J.; Gliwińska, A.; Badeński, A.; Szczepańska, M. Associations between renalase concentration and the occurrence of selected diseases. Endokrynol. Polska 2020, 71, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Qian, D.-W.; Jiang, S.; Shang, E.-X.; Zhu, Z.-H.; Duan, J.-A. Scutellariae Radix and Coptidis Rhizoma Improve Glucose and Lipid Metabolism in T2DM Rats via Regulation of the Metabolic Profiling and MAPK/PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2018, 19, 3634. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-Y.; Shi, C.-X.; Gao, R.; Sun, H.-J.; Xiong, X.-Q.; Ding, L.; Chen, Q.; Li, Y.-H.; Wang, J.-J.; Kang, Y.-M.; et al. Irisin inhibits hepatic gluconeogenesis and increases glycogen synthesis via the PI3K/Akt pathway in type 2 diabetic mice and hepatocytes. Clin. Sci. 2015, 129, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Hollander, L.; MacPherson, D.; Wang, L.; Velazquez, H.; Chang, J.; Safirstein, R.; Cha, C.; Gorelick, F.; Desir, G.V. Inhibition of renalase expression and signaling has antitumor activity in pancreatic cancer. Sci. Rep. 2016, 6, 22996. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Velazquez, H.; Moeckel, G.; Chang, J.; Ham, A.; Lee, H.T.; Safirstein, R.; Desir, G.V. Renalase Prevents AKI Independent of Amine Oxidase Activity. J. Am. Soc. Nephrol. 2014, 25, 1226–1235. [Google Scholar] [CrossRef]
- Tokinoya, K.; Shiromoto, J.; Sugasawa, T.; Yoshida, Y.; Aoki, K.; Nakagawa, Y.; Ohmori, H.; Takekoshi, K. Influence of acute exercise on renalase and its regulatory mechanism. Life Sci. 2018, 210, 235–242. [Google Scholar] [CrossRef]
- Barnard, R.; Faria, D.J.; Menges, J.E.; Martin, D.A. Effects of a high-fat, sucrose diet on serum insulin and related atherosclerotic risk factors in rats. Atherosclerosis 1993, 100, 229–236. [Google Scholar] [CrossRef]
- Barnard, R.J.; Roberts, C.K.; Varon, S.M.; Berger, J.J. Diet-induced insulin resistance precedes other aspects of the metabolic syndrome. J. Appl. Physiol. 1998, 84, 1311–1315. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Q.; Ma, W.; Tian, F.; Shen, H.; Zhou, M. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 2017, 8, 4644–4656. [Google Scholar] [CrossRef]
- Lu, L.; Tang, M.; Li, J.; Xie, Y.; Li, Y.; Xie, J.; Zhou, L.; Liu, Y.; Yu, X. Gut Microbiota and Serum Metabolic Signatures of High-Fat-Induced Bone Loss in Mice. Front. Cell. Infect. Microbiol. 2021, 11, 788576. [Google Scholar] [CrossRef]
- Yang, B.; Zheng, F.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Chen, W. Lactobacillus reuteri FYNLJ109L1 Attenuating Metabolic Syndrome in Mice via Gut Microbiota Modulation and Alleviating Inflammation. Foods 2021, 10, 2081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fang, R.; Lu, X.; Zhang, Y.; Yang, M.; Su, Y.; Jiang, Y.; Man, C. Lactobacillus reuteri J1 prevents obesity by altering the gut microbiota and regulating bile acid metabolism in obese mice. Food Funct. 2022, 13, 6688–6701. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Colonized Niche, Evolution and Function Signatures of Bifidobacterium pseudolongum within Bifidobacterial Genus. Foods 2021, 10, 2284. [Google Scholar] [CrossRef] [PubMed]
- Lieber, A.D.; Beier, U.H.; Xiao, H.; Wilkins, B.J.; Jiao, J.; Li, X.S.; Schugar, R.C.; Strauch, C.M.; Wang, Z.; Brown, J.M.; et al. Loss of HDAC6 alters gut microbiota and worsens obesity. FASEB J. 2018, 33, 1098–1109. [Google Scholar] [CrossRef]
- Yuan, G.; Tan, M.; Chen, X. Punicic acid ameliorates obesity and liver steatosis by regulating gut microbiota composition in mice. Food Funct. 2021, 12, 7897–7908. [Google Scholar] [CrossRef]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.C.H.; Wang, Y.-X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 2020, 70, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xu, J.; Sheng, Y.; Liu, J.; Li, H.; Guo, M.; Xu, W.; Luo, Y.; Huang, K.; He, X. Pleurotus ostreatus Ameliorates Obesity by Modulating the Gut Microbiota in Obese Mice Induced by High-Fat Diet. Nutrients 2022, 14, 1868. [Google Scholar] [CrossRef]
- Cheng, X.; Jiang, J.; Li, C.; Xue, C.; Kong, B.; Chang, Y.; Tang, Q. The compound enzymatic hydrolysate of Neoporphyra haitanensis improved hyperglycemia and regulated the gut microbiome in high-fat diet-fed mice. Food Funct. 2022, 13, 6777–6791. [Google Scholar] [CrossRef]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K.; et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18933–18938. [Google Scholar] [CrossRef]
- Fatima, S.S.; Jamil, Z.; Alam, F.; Malik, H.Z.; Madhani, S.I.; Ahmad, M.S.; Shabbir, T.; Rehmani, M.N.; Rabbani, A. Polymorphism of the renalase gene in gestational diabetes mellitus. Endocrine 2016, 55, 124–129. [Google Scholar] [CrossRef]




| Fatty Acids Name | Contents | Fatty Acids Name | Contents | Fatty Acids Name | Contents | Fatty Acids Name | Contents |
|---|---|---|---|---|---|---|---|
| C10, Capric | 0.1 | C16:1, Palmitoleic | 3.4 | C18:3, Linolenic | 5.2 | C20:4, Arachidonic | 0.7 |
| C12, Lauric | 0.2 | C17 | 0.9 | C20, Arachidic | 0.4 | C22:5, Docospentaenoic | 0.2 |
| C14, Myristic | 2.8 | C18, Stearic | 26.9 | C20:1 | 1.5 | ||
| C15 | 0.2 | C18:1, Oleic | 86.6 | C20:2 | 2.0 | ||
| C16, Palmitic | 49.9 | C18:2, Linoleic | 73.1 | C20:3 | 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, H.; Aoki, K.; Tokinoya, K.; Yonamine, M.; Sugasawa, T.; Kawakami, Y.; Takekoshi, K. Effects of High-Fat Diet on the Gut Microbiota of Renalase Gene Knockout Mice. Obesities 2022, 2, 303-316. https://doi.org/10.3390/obesities2030025
Fang H, Aoki K, Tokinoya K, Yonamine M, Sugasawa T, Kawakami Y, Takekoshi K. Effects of High-Fat Diet on the Gut Microbiota of Renalase Gene Knockout Mice. Obesities. 2022; 2(3):303-316. https://doi.org/10.3390/obesities2030025
Chicago/Turabian StyleFang, Hui, Kai Aoki, Katsuyuki Tokinoya, Masato Yonamine, Takehito Sugasawa, Yasushi Kawakami, and Kazuhiro Takekoshi. 2022. "Effects of High-Fat Diet on the Gut Microbiota of Renalase Gene Knockout Mice" Obesities 2, no. 3: 303-316. https://doi.org/10.3390/obesities2030025
APA StyleFang, H., Aoki, K., Tokinoya, K., Yonamine, M., Sugasawa, T., Kawakami, Y., & Takekoshi, K. (2022). Effects of High-Fat Diet on the Gut Microbiota of Renalase Gene Knockout Mice. Obesities, 2(3), 303-316. https://doi.org/10.3390/obesities2030025








