Abstract
Lichens are symbiotic partnerships between a filamentous fungus and a photosymbiotic “alga”. Studies show that lichens harbor endothallic fungi, but that some taxa have been difficult to isolate from the main filamentous thallus-forming fungus and other faster growing lichenicolous/endothallic fungi. Therefore, we aimed to develop and evaluate liquid yeast-enrichment strategies to (1) isolate lichen-associated yeasts in pure culture, and (2) determine the taxonomic placement and breadth of the diversity of culturable yeasts. Eighty-two lichen samples were collected and washed with distilled water, and healthy thalli were ground up and added to seven different yeast-enrichment broths. Yeast colonies were isolated in pure culture and identified using molecular techniques. Initial isolates were identified using BLASTn analysis, and a taxonomic refinement was completed using PhyML analysis. In total, 215 isolates were obtained. The most prevalently isolated ascomycetous yeasts were within the Dothideomycetes (Aureobasidium, Plowrightia, and Dothiora), while the most frequently isolated basidiomycetous yeasts belonged to the genera Curvibasidium, Sporobolomyces, and Tremella. The generic placements could not be determined for 17 isolates, and in total 25 novel species were recovered. The results of this research indicate that (1) lichen-associated yeasts are diverse, (2) employing liquid enrichment strategies is effective for isolating many of these, and (3) lichen thalli represent a valuable untapped reservoir of diverse and novel yeast species.
1. Introduction
Yeasts are fungi that have a predominant single cell growth state and appear primarily in both Ascomycota and Basidiomycota. Ascomycetous yeasts are important in human food production and alcohol fermentation, and the ascomycetous yeast Saccharomyces cervisiae was the first eukaryotic organism to have its genome completely sequenced in 1996 []. Basidiomycetous yeasts often associate with plants and benefit human agriculture due to their potential for biocontrol and bioremediation [], and ascomycetous yeast–insect associations have been well documented []. In contrast, lichen–yeast association studies have been conducted to a lesser extent even though lichens are ubiquitous and have been reported to be dominate the constituents of northern boreal forests and tundra biomes, as seen in []. Interestingly, recent research has shown that (1) lichens contain endothallic yeast taxa [,], (2) there are yeast-like fungal pathogens in lichens, particularly within the fungal class Tremellomycetes [], and (3) some lichens have external bacterial biofilms that may contain fungi []. Of particular interest are the diverse lichen-associated species within the genus Cyphobasidium (Cystobasidiomycetes) which appear to be widely distributed [] but difficult to obtain in pure culture.
Direct plating methods are useful when yeast cells are numerous []. However, isolating a diverse group of yeast taxa (likely in low number) from a lichen thallus which is primarily composed of a filamentous fungus and a photosymbiont, along with isolating other associated filamentous fungi, can be challenging. Liquid enrichment strategies can be employed to selectively recover yeasts while suppressing the growth of filamentous fungi []. Importantly, liquid enrichment strategies can be tailored to reflect the environmental stresses that lichen-associated yeasts will encounter in nature. For example, lichens have no mechanisms to actively regulate water content, so lichen thallus water availability constantly fluctuates []. Therefore, the use of liquid enrichment media that vary water availability, as well as the source of carbon, with selective agents could be one selective strategy for the isolation of lichen-associated yeasts.
Additional strategies to use in concert with liquid enrichment include an investigation of the potential usability of common lichen secondary metabolites as nutrient sources, and that of selective agents that could inhibit the main thallus-forming fungus but not lichen-associated yeasts. For example, lichens are known to produce at least 400 different acids [] and many produce terpenes as secondary metabolites []. Terpenes have antifungal properties []; however, some basidiomycetous yeasts have been demonstrated, in vitro, to use limonene vapor, a cyclic monoterpene, as a sole carbon source []. Differences in chitin content between yeasts and filamentous fungi also offer a good target for selective inhibition. Yeast cells have between 0.5–5% chitin, while filamentous fungi have up to 20% or more chitin distributed throughout the entire hyphal cell wall [,]. Therefore, Nikkomycin Z, a chitin synthase inhibitor, has the potential to be a good selective agent.
The molecular identification of both ascomycetous and basidiomycetous yeasts generally relies on ribosomal DNA regions. Traditionally, domains 1 and 2 of the large ribosomal subunit (LSU) are used to distinguish yeast species from each other []. In addition, the internal transcribed spacer region (ITS) 1 and ITS 2 are also useful for separating some yeast species, but not all. Therefore, species identification using both LSU domains and ITS regions is recommended [] since only about 3% of yeast species cannot be delimitated with both loci []. A DNA barcoding study of 9000 yeast isolates suggested the following taxonomic thresholds for yeast species delimitation: an ITS sequence similarity of 98.4% and LSU sequence similarity of 99.51%. The authors of the study also determined the genus level threshold to be 96.31% and 97.11% for ITS and LSU, respectively [].
The goals of our study were to (1) determine the effectiveness of liquid enrichment strategies for isolating lichen-associated yeasts, (2) test the use of a selective carbon source for the isolation of lichen-associated yeasts, and (3) examine the use of a selective agent (chitin synthase inhibitor) for the isolation of lichen-associated yeasts. We expected that (1) liquid enrichment strategies would isolate a different set of lichen-associated fungal taxa to direct plating strategies, (2) there would be some lichen-associated yeasts that could be isolated using terpenes as a sole carbon source, and (3) there would be a differential isolation of lichen-associated yeasts after exposure to a chitin synthase inhibitor. This research is important because lichen thalli may represent a valuable untapped culturable reservoir of diverse and novel yeast species.
2. Materials and Methods
2.1. Lichen Sample Collection and Molecular Identification
Eighty-two lichen samples were collected from Indiana, North Carolina, and Pennsylvania, from March through June 2022 (Figure 1, Table 1) for liquid enrichment strategies. These locations were chosen to collect a wide range of lichens from different habitats. The Indiana samples were collected from mixed hard-wood forest, the North Carolina samples were collected from conifer-dominated forests, and the Pennsylvania samples were collected in mountainous regions with a focus on lichen species that grew on rock surfaces. The genera of the lichen samples were identified by determining rDNA ITS/LSU sequence similarity. The molecular identification of lichen samples was completed using a modifed NaOH extraction protocol []. In short, a BB-sized amount (c.a. 2 × 2 mm) of lichen thallus tissue was ground in a 1.5 mL centrifuge tube containing 200 μL 0.5 M NaOH solution and centrifuged 16,873× g for 2 min, and 5 μL of the resulting supernatant was added to 495 μL 100 mM Tris-HCl buffered with NaOH to pH 8.5−8.9. All PCRs were completed on an Eppendorf Mastercycler Pro S thermocycler with a total reaction volume of 25 μL (12.5 μL 2× MyTaq Mix (Bioline), 1 μL of each 20 μM primer ITS-1F/ITS4 (ITS-1F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′ and ITS4: 5′-TCCTCCGCTTATTGATATGC-3′) [,] or 1 μL of each 20 μM primer LROR/LR6 (LROR: 5′-ACCCGCTGAACTTAAGC-3′ and LR6: 5′-CGCCAGTTCTGCTTACC-3′) [,], a 2 μL DNA template, and 8.5 μL DNA-free water). The thermal cycle parameters included initial denaturation at 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 20 or 30 s, 72 °C for 1 min, and a final extension step of 72 °C for 10 min. Gel electrophoresis (1% TAE agarose gel) and SYBR GelRed was used to verify the presence of a PCR product. Both the PCR purification and Sanger sequencing were performed by Genewiz (Plainfield, NJ, USA). The base-calls from the resulting sequences were reviewed by Sequencer 5.4.6, and the final sequence was compared against the NCBI database by BLASTn analysis []. All the lichen samples were deposited into the Purdue University Kriebel Herbarium (PUL) (Table 1).

Figure 1.
Examples of lichen species sampled in this study. (A) Parmotrema sp.; (B) Xanthoparmelia sp.; (C) Cladonia sp.; (D) Umbilicaria sp.; (E) Usnea sp.; (F) Cladonia sp.

Table 1.
Identified lichen samples utilized in this study based on NCBI BLASTn analysis.
2.2. Liquid Enrichment Strategies
All the collected samples were processed within 7 days. The samples were externally rinsed with distilled water to remove loose debris and hydrate the lichen thallus. Using sterilized metal dental tools, crustose and foliose lichen thalli were scrapped from above in order to grind only the top and inner tissue layers. Care was taken to avoid the underlying substrate. The fruticose and squamulose tissues were allowed to dry overnight, cut into small pieces, transferred to sterile plastic weigh boats, and ground into a fine powder. For all thallus types, a portion (BB-sized) of the ground tissue was transferred into a separate glass test tube containing 5 mL of the following: (1) an acidified yeast extract broth (3 g yeast extract, 3 g malt extract, 5 g peptone, 10 g dextrose, 10% citric acid, 250 mg chloramphenicol, 1 L distilled water), (2) a high-osmotic broth (yeast-nitrogen-base, 20% glucose, 0.1% yeast extract, 0.01% malt extract, 250 mg chloramphenicol, 1 L distilled water), (3) a xerotolerant yeast broth (5 g peptone, 10 g glucose, 1 g KH2PO4, 0.5 g MgSO4-7H2O, 0.002 dicholoran (1 mL of 0.2% solution in ethanol), 100 mg chloramphenicol, 222 mL glycerol, 778 mL distilled water), and (4) a nitrogen-limited broth (300 mg L−1 chloramphenicol, 11.7 g Bacto-yeast-carbon base, 400 mg cycloheximide, 310 mg thymine, 1 L distilled water) []. All media were autoclaved at the standard pressure and temperature for 20 min. After the base media cooled, chloramphenicol, dicholoran, and cycloheximide were added antiseptically to the appropriate base medium. Inoculated test tubes were used at room temperature and manually inverted 1× per day for 5 days. Next, under laminar flow hood, 200 µL of supernatant from each culture tube was lawn-plated onto acidified malt extract agar in 90 mm sterilized plastic Petri plates. The plates were sealed with parafilm, incubated at 18–20 °C under 12 h light/12 h dark, and monitored for yeast colonies for two weeks. All yeast colonies with different morphologies per plate were transferred to individual plates containing potato dextrose agar (PDA) to obtain pure cultures. For several of the lichens, sterile cotton swabs were lightly rubbed on the outside of cleaned thallus tissue and placed directly into each medium to evaluate a larger portion of the external surface of lichens.
2.3. Evaluation of Terpenes as a Selective Liquid Enrichment Strategy
Lichens are known to produce terpenes [], and soil basidiomycete yeast species from monoterpene rich environments have previously been isolated using monoterpenes as a sole carbon source []. This indicates the potential for yeasts with similar metabolic capabilities to inhabit lichen thalli. Therefore, 10 rock associated lichen samples were evaluated to determine if monoterpenes could be used as a selective agent for lichen-associated yeasts. The base medium consisted of a modified Spezieller Nährstoffarmer medium (1 g KNO3, 1 KH2PO4·7H2O, 0.5 g NaCl, 1 L distilled water). The base medium was autoclaved as above, and 2.5 mL was added to sterilized culture tubes containing BB-sized scraps of lichen thallus. The uncapped tubes were placed into a sterilize beaker which was placed inside a 1-gallon glass jar. In addition, 5 mL of Pelargonium graveolens essential oil (Better Homes & Gardens) was added to a separate sterilized culture tube and placed into a separate beaker within the 1 gallon glass jar. The glass jar was sealed, placed on a shake culture at 18–20 °C under an 8 h light/16 h dark cycle at 100 rpms for 7 days. As above, 200 µL of supernatant was lawn-plated onto malt extract agar supplemented with 250 mg L−1 chloramphenicol, sealed with parafilm, and processed as above. Pelargonium graveolens essential oil was chosen since it was reported, using HPLC, to contain several different monoterpenes, gamma-Eudesmol (sequiterpene), citronellyl formate (carboxylic acid), an isomenthane, and geranyl formate (formate ester) []. In addition, the essential oil was not directly added to the medium but to the head space as in [] to reduce the potential toxicity of the terpenes.
2.4. Evaluation of Nikkomycin Z as a Selective Inhibitor with Liquid Enrichment Strategy
Nikkomycin Z is a nucleoside di-peptide that acts as a chitin synthase inhibitor and is known to have differential rates of uptake in fungi []. Therefore, 10 branch-associated lichens were assayed to determine if this nucleoside di-peptide could be used to selectively isolate lichen-associated yeasts. The growth medium consisted of a potato dextrose broth supplemented with 0.25 mg/L chloramphenicol. Three concentrations of Nikkomycin Z were assayed per lichen (2 µg/mL, 8 µg/mL, and 14 µg/mL). For each assay, 5 mL of each broth was added to glass test tubes, and lichen tissue was obtained as previously described. The test tubes were incubated at 18–20 °C and manually inverted 1× per day for 5 days. Next, under a laminar flow hood, 200 µL of supernatant from each culture tube was lawn-plated onto PDA in 90 mm sterilized plastic Petri plates. The plates were sealed with parafilm, incubated at 18–20 °C under 12 h light/12 h dark, and monitored for yeast colonies for two weeks. All yeast colonies with different morphologies per plate were transferred to individual plates containing PDA to obtain pure cultures.
2.5. Direct Plating Methods
The growth medium consisted of the following: 6 g cornmeal agar (Difco, Moline, IL, USA), 8 g malt extract agar (Difco), 10 g PDA, 6 g yeast extract, 6 g peptone, 18 g oatmeal agar (Difco), 0.2 g CaCO2, 6 g agar, and 1 L distilled water. This medium was autoclaved at the standard temperature and pressure for 20 min and allowed to cool prior to the addition of 0.25 mg/L chloramphenicol. One set of lichen thalli were surface-sterilized by submerging the tissue in bleach solution (5% available chlorine) for 1 min. followed by rinsing them for 1 min. in autoclaved water two separate times. Another set of lichen thalli was processed as above without the bleach surface sterilization step. Next, each lichen tissue was placed into 1 mL of sterilized 0.9% saline solution and ground with a sterilized pestle. Finally, 100 µL of this solution was lawn-plated onto 90 mm Petri plates containing the above medium. The petri plates were wrapped with parafilm, incubated at 18–20 °C under a 8 h light/16 h dark cycle, and monitored every two days, for two weeks, for the growth of yeast colonies. All yeast isolates with unique morphologies per plate were transferred to separate Petri plates containing the same medium.
2.6. Fungal Isolate Identification, Phylogenetic Placement Analyse, Visualization, and Storage
Fungal isolate DNA was extracted by placing a BB-sized piece of the axenic colony into 500 µL of autoclaved milipore water, ground with a sterilized plastic pestle, and microwaved for 2 min. at 20 s intervals. Both the ITS/LSU sequencing and molecular identification was completed as in Section 2.1. In addition, all isolates with less than 98.4% of the ITS taxonomic threshold [] were further compared to sequences in the BOLDSYSTEMS fungal ITS database []. To further refine the identification of the isolates, the isolates were placed into phylogentic trees using scaffolding sequences from the NCBI database (Supplemental Table S1). In short, LSU sequences were aligned in SeaView 5.05 [] using Muscle []. A Gblocks analysis [] was completed with the following settings: (1) smaller final blocks, (2) gap positions within the final blocks, and (3) less strict flanking positions. In SeaView, a PhylML tree was constructed using the GTR model with the following settings: (1) 100 bootstrap replicates, (2) an optimized nucleotide equilibrium, (3) optimized invariable sites, (4) optimized rate variation across sites, (5) the best of the NNI and SPR tree searching operations, and (6) an optimized starting tree. The final resulting PhyML tree was visualized with metadata in Evolview 2 []. All cultures were transferred to PDA slants (stored at 4 °C) and 40% glycerin tubes (−80 °C) for long term storage.
3. Results
A total of 215 yeast and yeast-like isolates from at least 40 genera were isolated from 82 lichen samples using liquid enrichment and selective strategies (Table 2). The visual examination of the lichen thalli indicated the presence of external lichen-associated fungi (Figure 2A–C). The visualized fungi were attached to the external lichen thallus and grew in vitro via direct plating onto PDA (Figure 2B,E).

Table 2.
Generic diversity of culturable lichen-associated yeasts using liquid enrichment and selective strategies and direct plating.
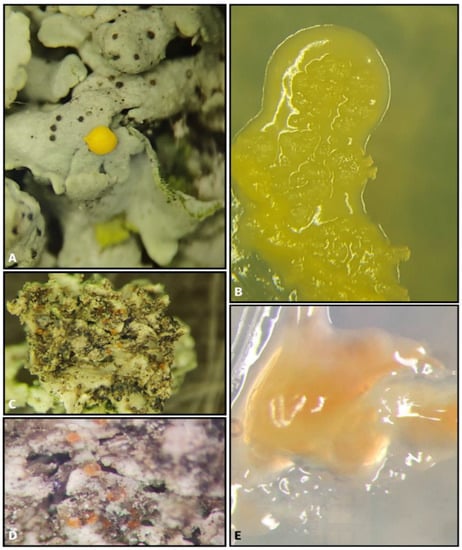
Figure 2.
Lichen-associated yeasts. (A) Tremellomycetes growing on upper surface of Physcia sp. thallus and (B) isolated on PDA; (C,D) Tremellomycetes growing on the under surface of Physcia sp. thalli and (E) isolated on PDA.
3.1. Taxonomy of Lichen-Associated Yeasts from Liquid Enrichment Strategies
Isolates from the phylum Ascomycota constituted 52% of the total number while isolates from the phylum Basidiomycota constituted the remainder. The Ascomycota isolates were from five fungal classes: Dothideomycetes (77%) (Figure 3 and Figure 4), Eurotiomycetes (5%), Leotiomycetes (4%), Saccharomycetes (5%), and Sordariomycetes (10%) (Figure 3 and Figure 4). The Ascomycota isolates consisted of 18 genera and 9 isolates of an undetermined genus (Table 2). The three Ascomycota genera with the most isolates were all within the Dothideomycetes (Aureobasidium (51%), Plowrightia (13%), and Dothiora (6%)) (Figure 4). The Basidiomycota isolates were found in the classes Cystobasidiomycetes (2%), Microbotryomycetes (46%), Pucciniomycetes (1%), and Tremellomycetes (51%) (Figure 5 and Figure 6). The Basidiomycetes isolates consisted of 24 genera and 8 isolates of an undetermined genus (Table 2). The three Basidiomycota genera with the most isolates were Curvibasidium (19%), Sporobolomyces (12%), and Tremella (12%) (Figure 5 and Figure 6).
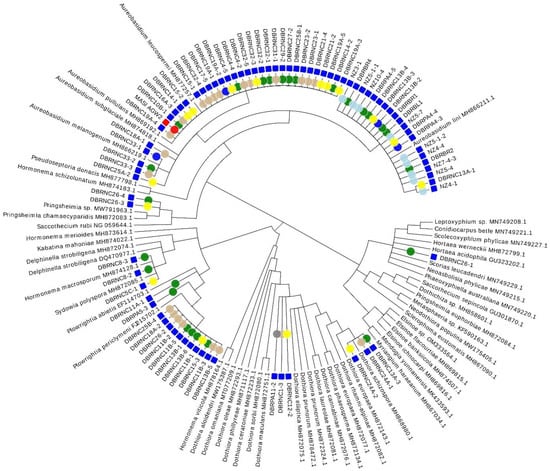
Figure 3.
Cladogram of cultured lichen-associated Dothideomycetes isolates showing the distribution of isolates obtained across different isolation media. Cladogram reconstructed from LSU alignment, GBLOCK analysis, and PhyML tree reconstruction. Visualization conducted using EvolView 2. Blue squares represent isolates obtained via liquid enrichment strategies; red square represents isolates obtained via direct plating methods; taxa with no information are class representative taxa. Circles represent isolation medium: red circle = direct plating; green circle = acidified malt broth; tan circle = xerophilic broth; blue circle = monoterpene; light blue = Nikkomycin Z; yellow = high osmotic broth; grey = saline swab.
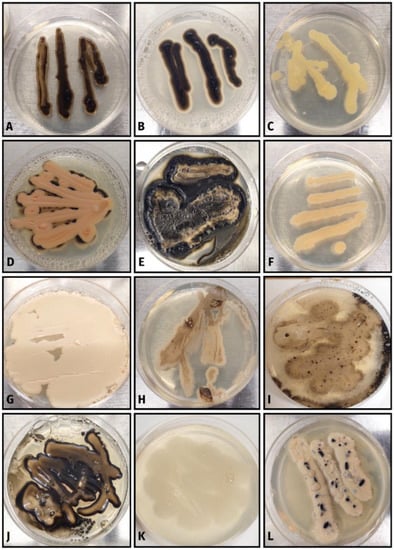
Figure 4.
Lichen-associated isolates from the class Dothideomycetes. (A–C) Dothiora isolates; (D,E) Plowrightia isolates; (F) unknown Dothideales isolate; (G–L) Aureobasidium isolates.
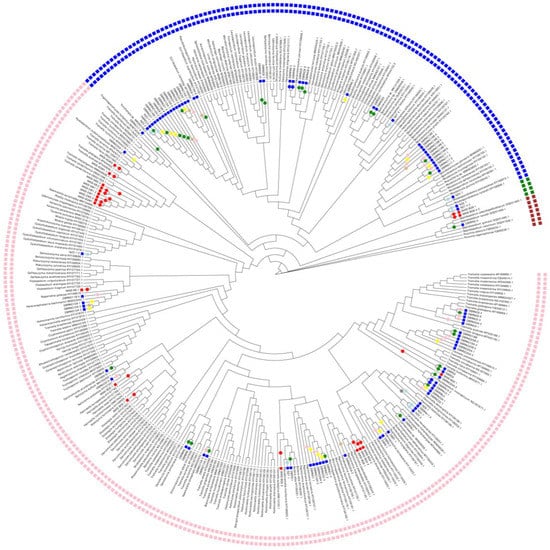
Figure 5.
Cladogram of cultured lichen-associated Cystobasidiomycetes, Microbotryomycetes, Pucciniomycetes, and Tremellomycetes isolates showing the distribution of isolates obtained across different isolation media. Cladogram reconstructed from LSU alignment, GBLOCK analysis, and PhyML tree reconstruction. Visualization conducted using EvolView 2. Inner blue squares represent isolates obtained via liquid enrichment strategies; inner red squares represent isolates obtained via direct plating methods; taxa with no information are class representative taxa. Branch circles represent isolation medium: red circle = direct plating; green circle = acidified malt broth; tan circle = xerophilic broth; blue circle = monoterpene; pink = nitrogen free; light blue = Nikkomycin Z; yellow = high osmotic broth; grey = saline swab. Outer squares: Pink = Tremellomycetes, Blue = Microbotryomycetes, Green = Cystobasidiomycetes, and dark red = Pucciniomycetes.
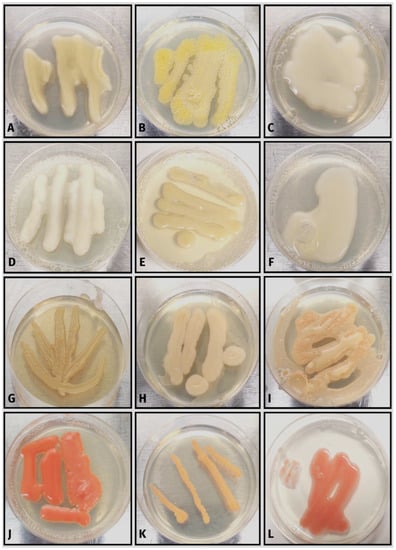
Figure 6.
Lichen-associated isolates from the Basidiomycota classes Microbotryomycetes and Tremellomycetes: (A–C) Tremella spp.; (D) Heterocephalacria sp.; (E) Kwoniella sp.; (F) Papiliotrema sp.; (G) Carcinomyces sp.; (H,I) Curvibasidium nothofagi; (J,K) Sporobolomyces spp.; (L) Rhodotorula sp.
3.2. Evaluation of Liquid Enrichment Strategies for the Isolation of Lichen-Associated Yeasts
There was little taxonomic overlap between the isolates obtained from the liquid enrichment assays when compared to that of previous isolates from the direct plating methods. Taphrina, Malassezia, and Erythrobasidium were some of the genera isolated solely by the direct plating methods. In contrast, the liquid enrichment assays isolated a broad range of fungal taxa. Two unknown Tremellomycetes isolates and two Phialophora isolates were only isolated using the nitrogen limited broth. Three unknown Tremellomycetes, one unknown Ascomycota isolate, and one unknown Dothideales isolate were obtained with the acidified yeast extract broth. One unknown Tremellomycetes and one unknown Dothideales isolate were obtained using the high osmotic broth. The xerotolerant yeast broth isolated a wide range of ascomycetous and basidiomycetous yeasts including unique Dothiora and Aureobasidium isolates and a unique Sporobolomyces isolate.
3.3. Evaluation of Terpenes and Nikkomycin Z for the Isolation of Lichen-Associated Yeasts
The basidiomycetous yeast genera isolated with P. graveolens essential oil as the sole carbon source included Curvibasidium, Fibulobasidium (presumptive), and Libkindia, while the ascomycetous yeasts included one unknown isolate within the Leotiomycetes genus (Supplemental Figure S1). The Nikkomycin Z, used as a selective agent, isolated 31 isolates (Figure 3 and Figure 5), of which four isolates were novel: two unknown Cystobasidiomycetes and two unknown Leotiomycetes isolates (Supplemental Figure S1). The fungal isolates were obtained from the entire Nikkomycin Z concentration range (2–14 µg/mL).
4. Discussion
The lichen thallus is known to house yeasts, both externally and internally, but many of these have not been cultured, as indicated by studies using culture-independent techniques [,]. In this study, we attempted a variety of different strategies for obtaining lichen-associated yeasts in culture. Fungal cultures are extremely important for obtaining fungal phenotypic, physiological, and ecological information, as well as in terms of their biomedical and industrial applications and are also necessary for formal species descriptions (e.g., []). Here, we used eight different isolation strategies on eighty-two lichen collections and recovered 215 isolates representing fifty-two species across twelve classes in two phyla. Of these, twenty-five are likely undescribed species based on the ITS and/or LSU sequence similarity against current taxa within the NCBI and BOLDSYSTEMS databases. The PhyML tree reconstruction, using LSU sequence data and reference sequences, also suggests that these twenty-five species are novel in addition to supporting three potentially undescribed genera. For several of the isolates, high quality LSU and ITS sequences were insufficient for placing species within their respective class or family. Therefore, multi-gene phylogeny will be required to determine their taxonomic placement.
There was little overlap in the isolated taxa when comparing the direct plating methods vs. the liquid enrichment strategies, and each enrichment strategy recovered different yeast species (Figure 3 and Figure 5). Despite the variety of methods employed, we were unable to obtain any Cyphobasidium species in culture, a genus known from sequencing data to be present in many lichens []. Therefore, we know that there are additional yeast taxa yet to be isolated. Future attempts should use liquid media infused with lichen extract, as shown by [], to isolate unique lichen-associated bacterial species. Lastly, several novel isolates were obtained using terpenes and Nikkomycin Z in conjunction with liquid enrichment strategies, suggesting that a further evaluation of these selective agents is warranted. Lastly, although it was not a direct goal of this study, it is important to understand the distribution of lichen-associated yeast species across hosts to determine which yeast species have host specificity or broad host distribution. For this study, fungal taxa from host lichen thalli can be found in Supplemental Table S2. Future research should focus on employing these methods to investigate the distribution of lichen-associated yeasts across lichen species.
4.1. Lichen-Associated Ascomycetous Yeast
Predominant lichen-associated Ascomycota classes have been previously reported to be Dothideomycetes, Eurotiomycetes, Leotiomycetes, and Sordariomycetes []. Our results are similar to an ITS1 metabarcoding study by [] which also obtained Dothideomycetes, Eurotiomycetes, and Leotiomycetes sequence representatives from rock-associated lichens. In our study, taxa from the Dothideomycetes family Dothioraceae were isolated, while [] reported that the most representative families within Dothideomycetes were Mycosphaerellaceae, Myriangiaceae, and Teratosphaeriaceae. For the Eurotiomycetes, [] taxa within Herpotrichiellaceae were prominent in []. We also obtained several representative isolates from this family. These differences are not surprising since environmental sampling and culturing techniques are considered complementary [] due to (1) method biases, and (2) because most fungal taxa are still considered unculturable. Additionally, we focused solely on lichen-associated yeast, while [] focused on the entire mycobiome. In the following section, we review the Ascomycota and Basidiomycota isolates we obtained, starting with the Dothideomycetes and the Leotiomycetes. The Dothideomycetes isolates are reviewed because this class contained 77% of the total ascomycetous yeast isolates, and the Leotiomycetes yeast and yeast-like isolates are discussed due to the low prevalence of their isolation.
Most ascomycetous lichen-associated yeasts in this study were within the Dothideomycetes class, particularly within the genera Aureobasidium, Dothiora, and Plowrightia (Figure 3 and Figure 4). Aureobasidium pullulans has been previously isolated from bark-associated lichen thalli [] and from rock-associated lichen thalli []. Similarly, in this study, we isolated several Aureobasidium species from both bark-associated and rock-associated lichen species (Figure 4G–L and Figure 5). Aureobasidium species have also been isolated directly from rock surfaces []. The authors of [] obtained a high proportion of ascomycetous yeasts from their sampled lichens, with both Candida sphagnicola and yeast isolates from the genus Dothiora being predominant. In addition, the authors of [] found Dothiora species to be localized in the upper actively growing thallus. In our study, the genus Dothiora represented only 6% of the isolated ascomycetous yeasts. The Dothiora isolates varied in pigmentation from dark-pigmented to a light-yellow color (Figure 4A–C). The two dark pigmented isolates clustered with D. schizospora, while the light-yellow-colored isolate appears to be a novel isolate within the genus Dothiora (Figure 3). Ecologically, Dothiora species are thought to be saprobic or weakly pathogenic []. As of 2018, Plowrightia mereschkowskyi appears to be the only Plowrightia species previously reported to be isolated from lichen thalli [,]. This Plowrightia species was isolated from Aspicilia hispida and is reported to be and obligate lichenicolous species [,]. Interestingly, we isolated a total of 13 isolates, representing three unique taxa (Figure 4D,E) based on phylogenetic analysis (Figure 3), from 6 different lichen genera: Aspicilia, Lecanora, Parmotrema, Physcia, and Usnea. Our results for lichen-associated Plowrightia species provide new host and new geographical connections.
Within the Leotiomycetes, we found three presumptive species, with only one being identified to the genus Tricellula (Supplemental Figure S1A). We note that the Tricellula isolate was more yeast-like upon initial isolation but converted to a slow-growing mycelial colony after one week on PDA. The other two yeast species appear to fall within the order Lichnodiales (Supplemental Figure S1A) based on [], and these isolates continued to produce yeast cells on a solid PDA medium (Supplemental Figure S1B–E). Based on our phylogenetic placement, isolate DBRPA9-1 is close to Mycosymbioces; however, the LSU sequence similarity between DBRPA9-1 and M. mycenophila is only 95%. Taxa within Lichnodiales are known to be lichenicolous [], and M. mycenophila is a mycoparasite [,], suggesting that at least one or more of these species could be mycoparasites of lichens, but further research would be required to determine the ecological roles of these isolates. Yeast and yeast-like morphologies in the Leotiomycetes are infrequently reported but do exist particularly within the orders Leotiales, Phacidiales, and Thelebolales [].
4.2. Lichen-Associated Basidiomycetous Yeast
Interest in lichen-associated yeasts, particularly within Cystobasidiomycetes, has grown since the discovery of endothallic basidiomycetous yeasts (genus Cyphobasidium) in a wide range of lichen species []. Based on this discovery, it was hypothesized that these taxa may represent a third obligate member of lichen symbiosis [], although additional research is needed to support this hypothesis. Currently, no Cyphobasidium species have been obtained in pure culture. However, researchers have discovered many lichen-associated Cystobasidiomycetes [,] and other basidiomycetous yeasts across many lichen taxa using both culture-dependent and culture-independent methods. In this study, we observed basidiomycetous yeast species from four fungal classes using liquid enrichment strategies and two selective agents (terpenes and Nikkomycin Z) (Table 2). Both Cystobasidiomycetes and Pucciniomycetes were represented by one species each, while Microbotryomycetes and Tremellomycetes contained 97% of the total basidiomycetous yeast isolates.
Cystobasidiomycetes species have diverse ecological roles, including in their roles as parasites, saprobes, and inhabitants of the phylloplane []. We isolated one unknown Cystobasidiomycetes species from two different lichen thalli (Candelaria concolor and one unidentified lichen species). Based on the NCBI blast match of the ITS and LSU rDNA regions, it appears to be within the genus Buckleyzyma, but further analysis is required for species level placement. We also isolated a Septobasidium species. Septobasidium species are emtomopathogens of scale insects [], so it is unlikely that this isolate is directly associated with lichen.
Microbotryomycetes species are mostly parasites of other fungi, parasites of plants, or saprotrophs []. In this study, the genera Curvibasidium and Sporobolomyces were the most common. Other researchers have isolated Curvibasidium rogersii and C. cygneicollum from Usnea [,], while we isolated C. nothofagi from nine separate samples consisting of C. concolor, Lecanora sp., Parmotrema sp., Porpidia sp., and Usnea species (Figure 6H,I). Sporobolomyces spp. in this study were isolated mainly with a nitrogen-free medium and a high-osmotic medium (Figure 5) and varied in colony color (Figure 6J,K). Using next generation sequencing techniques, Sporobolomyces has been previously identified from Hypogymnia hypotrypa [], and S. salmonicolor was previously isolated from a soil lichen [].
Tremellomycetes species are considered one of the predominant taxa isolated from lichen thalli []. For example, the ITS1 metabarcoding study by the authors of [] found that most sequence reads were associated with the class Tremellomycetes, in the order Tremellales, and within the genus Fibulobasidium. In this study, Tremellomycetes isolates were the most abundant, with Tremella spp. being prominent, but only a few Fibulobasidium isolates were obtained. The isolates varied in color from shades of cream, yellow, or orange. In addition, the Tremella taxa that were isolated using the direct plating method were distinct from those taxa isolated using the liquid enrichment and selective agent strategies (Figure 5). Ecologically, Tremellomycetes species are saprotrophs and mycoparasites, and some taxa are considered fungicolous [].
4.3. Methodological Observations and Considerations
The use of liquid enrichment strategies is not unique to isolating fungi from environmental sources. For example, the authors of [] used liquid enrichment to isolate fungi from marine sediment, the authors of [] used liquid enrichment to isolate benzene-degrading yeasts from acidic soils, and the authors of [] used liquid enrichment to isolate hydrocarbon-degrading black yeasts. In this study, we used several liquid enrichment media with the sole purpose of preventing the growth of the main lichen-forming fungus while allowing the lichen-associated yeasts to grow. Unfortunately, we observed that for some lichen species the major thallus-forming fungus was able to grow; therefore, further refinement and evaluation of additional selective media is ongoing. Although Nikkomycin Z appears to be promising, the current high cost of Nikkomycin Z makes it prohibited to be incorporated into routine isolation practices.
Although we did not directly measure media oxygen levels, we did observe spherical multipolar budding yeasts cells (Supplemental Figure S2) in liquid culture that were identified to the genus Mucor. For some dimorphic Mucor species, the yeast state is generally formed in low-oxygen environments, while the Mucor mycelium state is typically formed in oxygenated environments [] (Supplemental Figure S2). Our Mucor isolates reverted to the mycelial state as soon as the yeast cells were transferred to solid PDA plates, so they were excluded. However, this observation does suggest that our liquid enrichment methods produced a lower oxygen level compared to the solid plate medium. This could potentially limit the isolation of yeasts that require atmospheric oxygen levels. Therefore, future isolations will utilize once-a-day mixing and a continuous shake culture to maximize yeast isolation. Although these liquid enrichment strategies have the potential to isolate dimorphic fungal taxa (not the desired taxa), it is of great importance to document dimorphic fungal taxa because many human fungal pathogens are dimorphic [].
We also note that the direct plating methods were previously conducted prior to performing the liquid enrichment strategies. Therefore, future analyses will compare direct plating and liquid enrichment strategies at the same time and from the same lichen thallus to limit the potential variability across thalli of the same species. Lastly, we recognize that lichen thalli can contain a broad group of fungal taxa, which are known to be from other stressful habitats, including human pathogens and rock-inhabiting fungi []. Therefore, it is important to note that our study took a broad definition of lichen-associated fungi which includes external, internal, and opportunistic environmental fungi that have the potential to associate with lichen tissue. Additional lichen species-specific sampling and analysis will be needed to determine the type of and the extent of these lichen-associations.
5. Conclusions
Here, we demonstrate the usefulness of a liquid enrichment strategy with respect to isolating lichen-associated yeasts. Using these techniques, we have isolated a wide taxonomic range of yeasts from Ascomycota and Basidiomycota. In addition, we visually documented the yeast-like state of a dimorphic Mucor species and isolated several putative new species and genera. The future implementation of these methods will be of great value for researchers interested in culturing a greater proportion of lichen-associated yeasts. Importantly, obtaining cultures will allow researchers to learn more about the diversity and ecologies of these individual species and provide insight into their role and importance within lichen thalli.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ecologies4010012/s1. Table S1: Fungal species acquired from NCBI GenBank used for phylogenetic placement; Figure S1: Unknown yeast and yeast-like lichen-associated isolates from the Ascomycota class Leotiomycetes; Table S2: Distribution of fungal isolates across sampled lichens; Figure S2: Dimorphic Mucor isolate obtained from liquid enrichment strategies.
Author Contributions
Conceptualization, D.B.R. and M.C.A.; methodology, D.B.R. and M.C.A.; validation, D.B.R. and M.C.A.; formal analysis, D.B.R. and M.C.A.; investigation, D.B.R. and M.C.A.; resources, M.C.A.; data curation, D.B.R. and M.C.A.; writing—original draft preparation, D.B.R. and M.C.A.; writing—review and editing, D.B.R. and M.C.A.; visualization, D.B.R. and M.C.A.; supervision, M.C.A.; project administration, M.C.A.; funding acquisition, M.C.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the USDA National Institute of Food and Agriculture Hatch, project 1010662 and US National Science Foundation DEB-2018098.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All high-quality sequence data have been uploaded to NCBI (lichens = OP970266–OP970297, fungal isolates = OP967194–OP967384). All lichen samples were deposited into the Purdue University Kriebel Herbarium.
Acknowledgments
We thank the members of the Aime lab and Haiden Nguyen for sample collection support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, J.M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.K.; Johri, B.N. Basidiomycetous yeasts: Current status. In Yeast Biotechnology: Diversity and Applications; Satyanarayana, T., Kunze, G., Eds.; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar] [CrossRef]
- Stefanini, I. Yeast-insect associations: It takes guts. Yeast 2018, 35, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Petzold, D.E.; Mulhern, T. Vegetation distributions along lichen-dominated slopes of opposing aspect in the Eastern Canadian subarctic. Arctic 1987, 40, 221–224. [Google Scholar] [CrossRef]
- Millanes, A.M.; Diederich, P.; Wedin, M. Cyphobasidium gen. nov., a new lichen inhabiting lineage in the Cystobasidiomycetes (Pucciniomycotina, Basidiomycota, Fungi). Fungal Biol. 2016, 120, 1468–1477. [Google Scholar] [CrossRef]
- Spribille, T.; Tuovinen, V.; Res, P.; Vanderpool, D.; Wolinski, H.; Aime, M.C.; Schneider, K.; Stabentheiner, E.; Toome, H.M.; Thor, G.; et al. Basidiomycete yeasts in the cortex of ascomycete mac-rolichens. Science 2016, 353, 488–492. [Google Scholar] [CrossRef]
- Millanes, A.; Diederich, P.; Ekman, S.; Wedin, M. Phylogeny and character evolution in jelly fungi (Tremellomycetes, Basid-iomycota, Fungi). Mol. Phylogenet. Evol. 2011, 61, 12–28. [Google Scholar] [CrossRef]
- Spribille, T. Relative symbiont input and the lichen symbiotic outcome. Curr. Opin. Plant Biol. 2018, 44, 57–63. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Fell, J.W.; Teun, B.T.; Vincent, R.V. Chapter 7 Methods for isolation, phenotypic characterization and maintenance of yeasts. In The Yeasts, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: New York, NY, USA, 2011; pp. 87–110. [Google Scholar]
- Gasulla, F.; Del Campo, E.M.; Casano, L.M.; Guéra, A. Advances in understanding of desiccation tolerance of lichens and li-chen-forming algae. Plants 2021, 10, 807. [Google Scholar] [CrossRef]
- Takahagi, T.; Endo, T.; Yamamoto, Y.; Sato, F. Lichen photobionts show tolerance against lichen acids produced by lichen mycobionts. Biosci. Biotechnol. Biochem. 2008, 72, 3122–3127. [Google Scholar] [CrossRef]
- Culberson, C.F. Chemical and Botanical Guide to Lichen Products; University of North Carolina Press: Chapel Hill, CA, USA, 1969; pp. 58–62. [Google Scholar]
- Kahriman, N.; Yazici, K.; Arslan, T.; Aslan, A.; Karaoglu, S.A.; Yayli, N. Chemical composition and antimicrobial activity of the essential oils from Evernia prunastri (L.) ach. and Evernia divaricata (L.) ach. Asian J. Chem. 2011, 23, 1937–1939. [Google Scholar]
- Thanh, V.N.; Smit, M.S.; Moleleki, N.; Fell, J.W. Rhodotorula cycloclastica sp. nov., Rodotorula retinophila sp. nov., and Rodotorula terpenoidalis sp. nov., three limonene-utilizing yeasts isolated from soil. FEMS Yeast Res. 2004, 4, 857–863. [Google Scholar]
- Chaffin, W.L.; Lopez-Ribot, J.L.; Casanova, M.; Gozalbo, D.; Martinez, J.P. Cell wall and secreted proteins of Candida albicans: Identification, function, and expression. Microbiol. Mol. Biol. Rev. 1998, 62, 130–180. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J. Fungal Cell Wall: Structure, Synthesis and Assembly; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Scorzetti, G.; Fell, J.W.; Fonseca, A.; Statzell-Tallman, A. Systematics of basidiomycetous yeasts: A comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002, 2, 495–517. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vries, M.; Verkleij, G.J.M.; Crous, P.W.; Boekhout, T.; et al. DN barcoding analysis of more than 9000 yeasts isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Osmundson, T.W.; Eyre, C.A.; Hayden, K.M.; Dhillon, J.; Garbelotto, M.M. Back to basics: An evaluation of NaOH and alternative rapid DNA extraction protocols for DNA barcoding, genotyping, and disease diagnostics from fungal and oomycete samples. Mol. Ecol. Resour. 2013, 13, 66–74. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of my-corrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, N., Gelfand, D., Sninsky, J., White, T., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Rehner, S.A.; Samuels, G.J. Molecular systematics of the Hypocreales: A teleomorph gene phylogeny and the status of their anamorphs. Can. J. Bot. 1995, 73, S816–S823. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Demarne, F.-E. ‘Rose-scented geranium’ a Pelargonium grown for the perfume industry. In Geranium and Pelargonium: The Genera Geranium and Pelargonium; Lis-Balchin, M., Ed.; Taylor & Francis: New York, NY, USA, 2002; pp. 193–211. [Google Scholar]
- Tariq, V.; Devlin, P. Sensitivity of fungi to Nikkomycin Z. Fungal Genet. Biol. 1996, 20, 4–11. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The barcode of life data system (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Gouy, M.; Tannier, E.; Comte, N.; Parsons, D.P. Seaview version 5: A multiplatform software for multiple sequence alignment, molecular phylogenetic analyses, and tree reconciliation. In Multiple Sequence Alignment; Methods in Molecular Biology; Katoh, K., Ed.; Humana: New York, NY, USA, 2021; pp. 241–260. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wei, X.L.; Zhang, Y.Q.; Liu, H.Y.; Yu, L.Y. Diversity and distribution of lichen-associated fungi in the Ny-Ålesund Region (Svalbard, High Arctic) as revealed by 454 pyrosequencing. Sci. Rep. 2015, 5, 14850. [Google Scholar] [CrossRef] [PubMed]
- Banchi, E.; Stankovic, D.; Fernández-Mendoza, F.; Gionechetti, F.; Pallavicini, A.; Muggia, L. ITS2 metabarcoding analysis complements lichen mycobiome diversity data. Mycol. Prog. 2018, 17, 1049–1066. [Google Scholar] [CrossRef]
- Aime, M.C.; Miller, A.; Aoki, T.; Bensch, K.; Cai, L.; Crous, P.W.; Hawksworth, D.L.; Hyde, K.; Kirk, P.; Lücking, R.; et al. How to publish a new fungal species, or name, version 3.0. IMA Fungus 2021, 12, 11. [Google Scholar] [CrossRef]
- Biosca, E.G.; Flores, R.; Santander, R.D.; Díez-Gil, J.L.; Barreno, E. Innovative approaches using lichen enriched media to improve isolation and capturability of lichen associated bacteria. PLoS ONE 2016, 11, e0160328. [Google Scholar] [CrossRef]
- Muggia, L.; Grube, M. Fungal diversity in lichens: From extremotolerance to interactions with algae. Life 2018, 8, 15. [Google Scholar] [CrossRef]
- Fernámdez-Mendoza, F.; Fleischhacker, A.; Kopun, T.; Grube, M.; Muggia, L. ITS1 metabarcoding highlights low specificity of lichen mycobiomes at a local scale. Mol. Ecol. 2017, 26, 4811–4830. [Google Scholar] [CrossRef] [PubMed]
- Romão, D.; Staley, C.; Ferreira, F.; Rodrigues, R.; Sabino, R.; Veríssimo, C.; Wang, P.; Sadowsky, M.; Brandão, J. Next-generation sequencing and culture-based techniques offer complementary insights into fungi and prokaryotes in beach sands. Mar. Pollut. Bull. 2017, 119, 351–358. [Google Scholar] [CrossRef]
- Suryanarayanan, T.S.; Govindarajulu, M.B.; Rajamani, T.; Tripathi, M.; Joshi, Y. Endolichenic fungi in lichens of Champawat district, Uttarakhand, northern India. Mycol. Progress 2017, 16, 205–211. [Google Scholar] [CrossRef]
- Selbmann, L.; Grube, M.; Onofri, S.; Isola, D.; Zucconi, L. Antarctic epilithic lichens as niches for black meristematic fungi. Biology 2013, 2, 784–797. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, G.S.; Zalar, P.; Urzì, C.; de Leo, F.; Yurlova, N.A.; Sterflinger, K. Relationships of dothideaceous black yeasts and meristematic fungi based on 5.8S and ITS2 rDNA sequence comparison. Stud. Mycol. 1999, 43, 31–37. [Google Scholar]
- Kachalkin, A.V.; Glushakova, A.M.; Pankratov, T.A. Yeast population of the Kindo Peninsula lichens. Microbiology 2017, 86, 786–792. [Google Scholar] [CrossRef]
- Crous, P.W.; Groenewald, J.Z. The genera of fungi—G 4: Camarosporium and Dothiora. IMA Fungus 2017, 8, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Clauzade, G.; Diederich, P.P.; Roux, C. Nelikenigintaj fungoj likenlogaj–Ilustrita determinlibro. Bull. Soc. Linn. Provence 1989, 1, 1–142. [Google Scholar]
- Diederish, P.; Lawrey, J.D.; Damien, E. The 2018 classification and checklist of lichenicolous fungi, with 2000 non-lichenized, obligately lichenicolous taxa. Bryologist 2018, 121, 340–425. [Google Scholar] [CrossRef]
- Ekanayaka, A.H.; Hyde, K.D.; Gentekaki, E.; McKenzie, E.H.C.; Zhao, Q.; Bulgakov, T.S.; Camporesi, E. Preliminary classification of Leotiomycetes. Mycosphere 2019, 10, 310–489. [Google Scholar] [CrossRef]
- Frank, J.L. Mycosymbioces mycenophila. Index Fungorum. 2014. Available online: https://www.gbif.org/species/9746710 (accessed on 1 November 2022).
- Edwards, A.; Leech, T.; Senior, I. A gall-inducing infection of Lepista spp. In Norfolk by Mycosymbioces mycenophila—Fist record for Britain. Field Mycol. 2020, 21, 119–123. [Google Scholar] [CrossRef]
- Tanney, J.B.; Quijada, L. Comments on the occurrence of yeast-like morphologies in the Leotiomycetes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005141. [Google Scholar] [CrossRef]
- Černajová, I.; Škaloud, P. The first survey of Cystobasidiomycetes yeasts in the lichen genus Cladonia; with the description of Lichenozyma pisutiana gen. nov., sp. nov. Fungal Biol. 2019, 123, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Cometto, A.; Leavitt, S.D.; Millanes, A.M.; Wedin, M.; Grube, M.; Muggia, L. The yeast lichenosphere: High diversity of ba-sidiomycetes from the lichens Tephromela atra and Rhizoplaca melanophthalma. Fungal Biol. 2022, 126, 587–608. [Google Scholar] [CrossRef] [PubMed]
- Aime, M.C.; Toome, M.; McLaughlin, D.J. 10 Pucciniomycotina. In Systematics and Evolution; McLaughlin, D., Spatafora, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 7A. [Google Scholar] [CrossRef]
- Petch, T. Note on the biology of the genus Septobasidium. Ann. Bot. 1911, 25, 842. [Google Scholar] [CrossRef]
- Oberwinkler, F. Yeasts in Pucciniomycotina. Myco. Prog. 2017, 16, 831–856. [Google Scholar] [CrossRef]
- Bai, L.; Chong, A.; Zhu, Y.; Wang, X.; Wang, M.; Li, J.; Zhang, Z.; Zhao, X. First isolation and identification of cold adaptive yeast Curvibasidium rogersii from Usnea lichen and genome-based studies of its biological properties. Acta Microbiol. Sin. 2022, 62, 567–578. [Google Scholar]
- Wang, Q.; Li, J.; Yang, J.; Zou, Y.; Zhao, X.-Q. Diversity of endophytic bacterial and fungal microbiota associated with the medicinal lichen Usnea longissimi at high altitudes. Front. Microbiol. 2022, 13, 958917. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, Y.; Wang, X.; Wei, X.; Wei, J. Lichen-associated fungal community in Hypogymnia hypotrypa (Parmeliaceae, Ascomycota) affected by geographic distribution and altitude. Front. Microbiol. 2016, 7, 1231. [Google Scholar] [CrossRef]
- Dimitrova, S.; Pavlova, K.; Lukanov, L.; Korotkova, E.; Petrova, E.; Zagorchev, P.; Kuncheva, M. Production of metabolites with antioxidant and emulsifying properties by Antarctic strain Sporobolomyces salmonicolor AL1. Appl. Biochem. Biotechnol. 2013, 169, 301–311. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; Dayo-Owoyemi, I.; Nobre, F.S.; Pagnocca, F.C.; Chaud, L.C.S.; Pessoa, A.; Felipe, M.G.A.; Sette, L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 2013, 17, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Middelhoven, W.J.; Koorevaar, M.; Schuur, G.W. Degradation of benzene compounds by yeasts in acidic soils. Plant Soil 1992, 145, 37–43. [Google Scholar] [CrossRef]
- Isola, D.; Scano, A.; Orrù, G.; Prenafeta-Boldú, F.X. Hydrocarbon-contaminated sites; is there something more than Exophiala xenobiotica? New insights into black fungal diversity using the long cold incubation method. J. Fungi 2021, 7, 817. [Google Scholar] [CrossRef] [PubMed]
- Deacon, J. Fungal Biology; Blackwell: Malden, MA, USA, 2006; p. 371. [Google Scholar]
- Muggia, L.; Fleischhacker, A.; Kopun, T.; Grube, M. Extremotolerant fungi from alpine rock lichens and their phylogenetic relationships. Fungal Divers. 2016, 76, 119–142. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).