Dominant Species-Physiognomy-Ecological (DSPE) System for the Classification of Plant Ecological Communities from Remote Sensing Images
Abstract
:1. Introduction
2. Materials and Methods
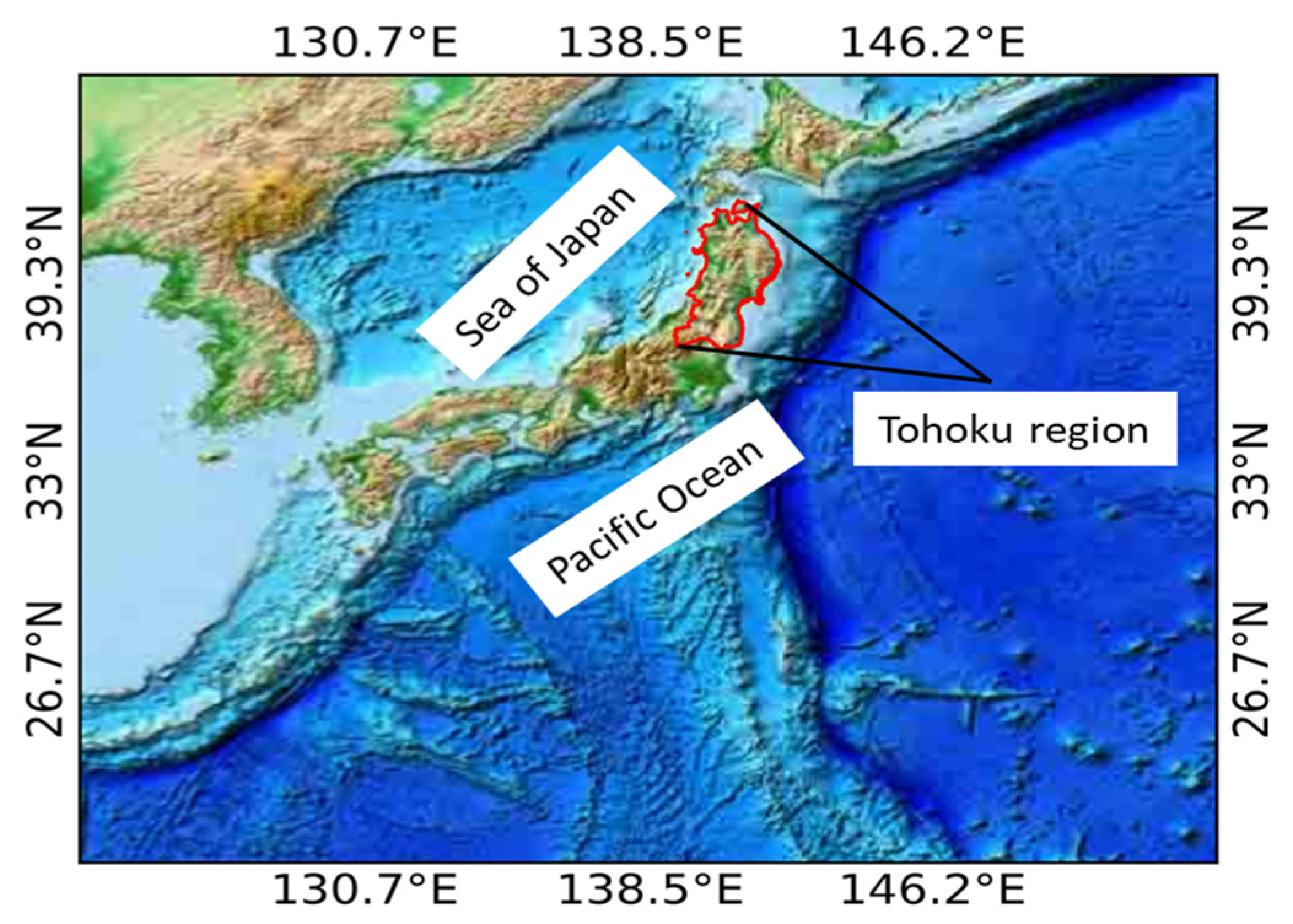
2.1. Study Area
2.2. DSPE Classification System
2.3. Preparation of Ground Truth Data
2.4. Processing of Satellite Data
2.5. Deep Recurrent Learning
3. Results
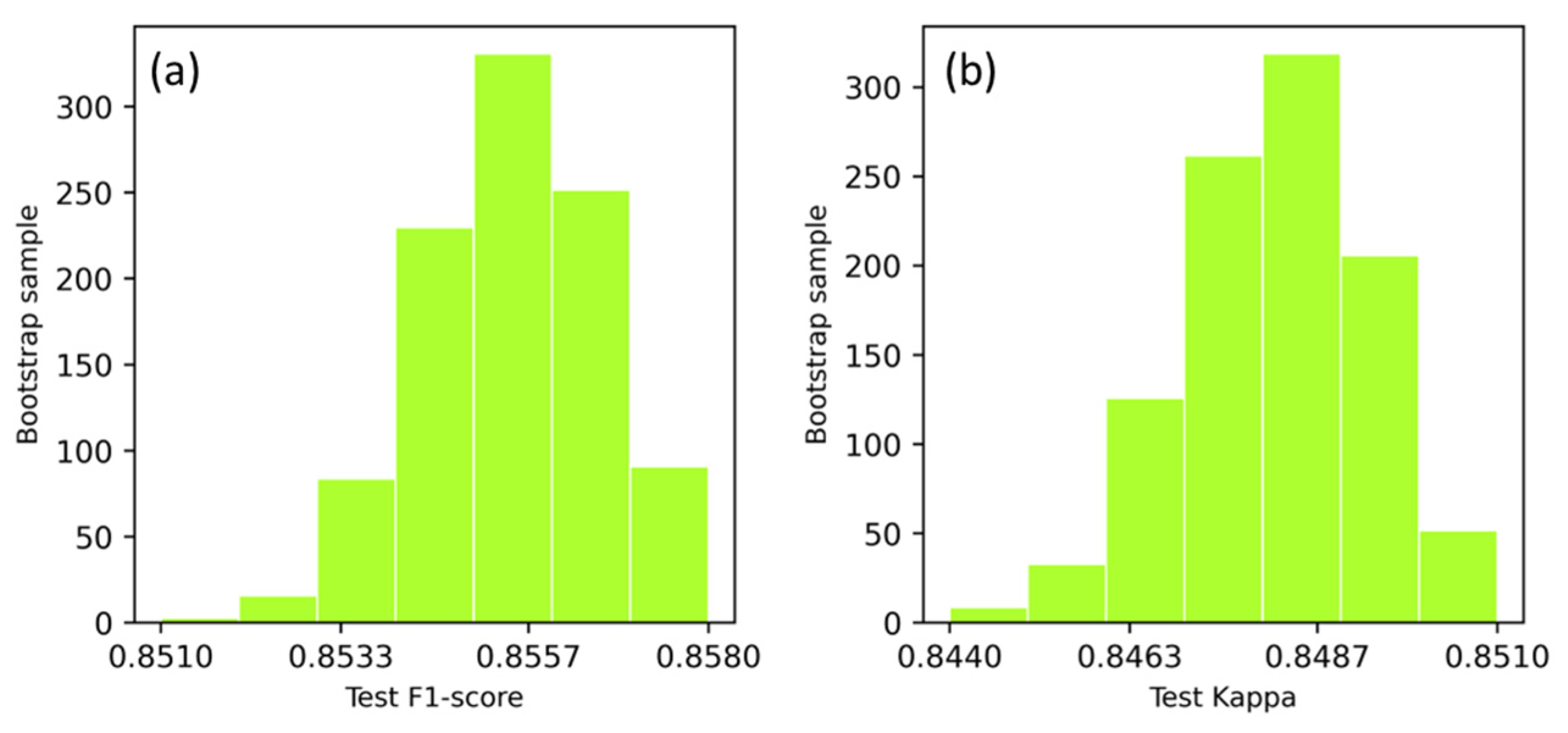
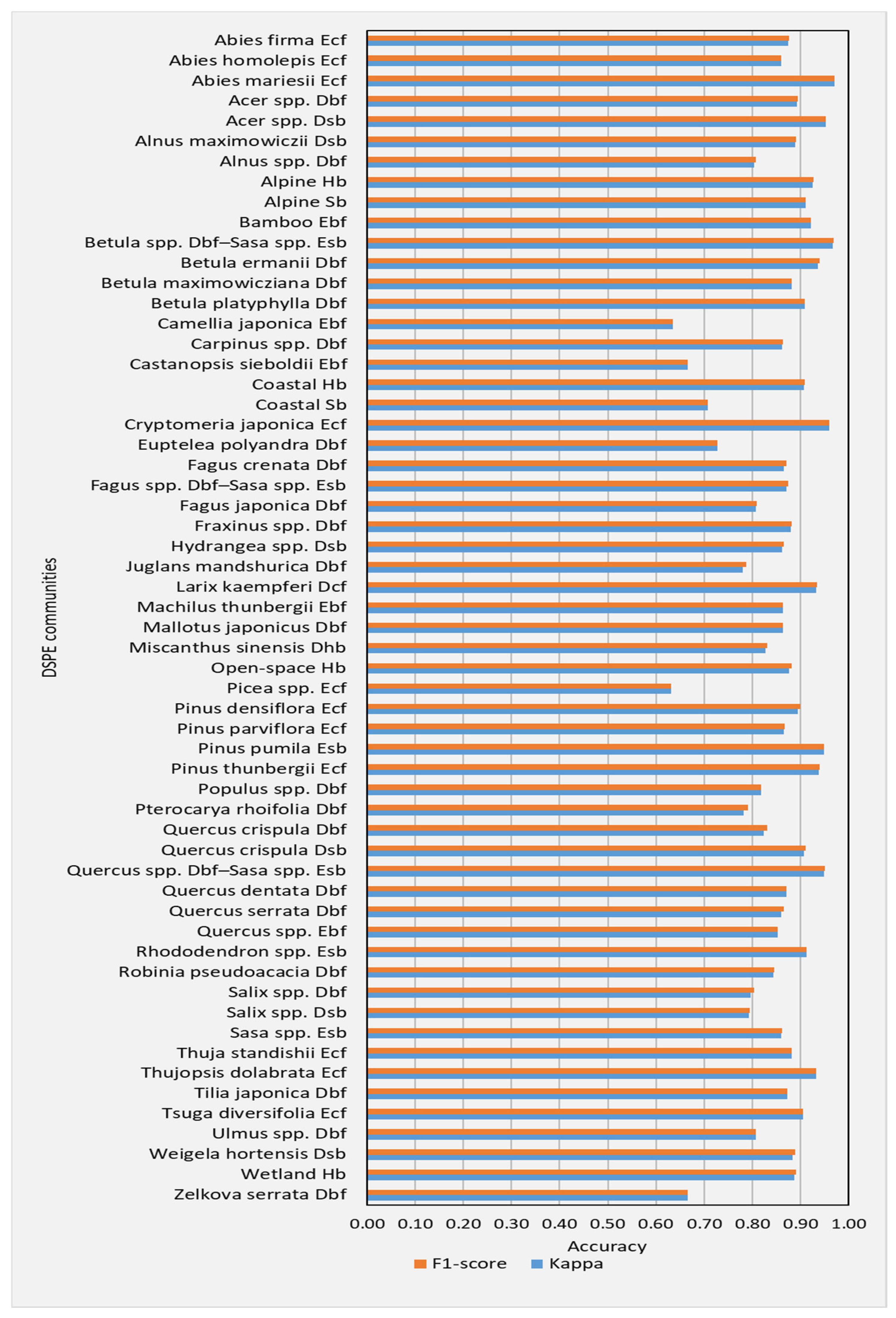
3.1. Performance of DSPE Classification
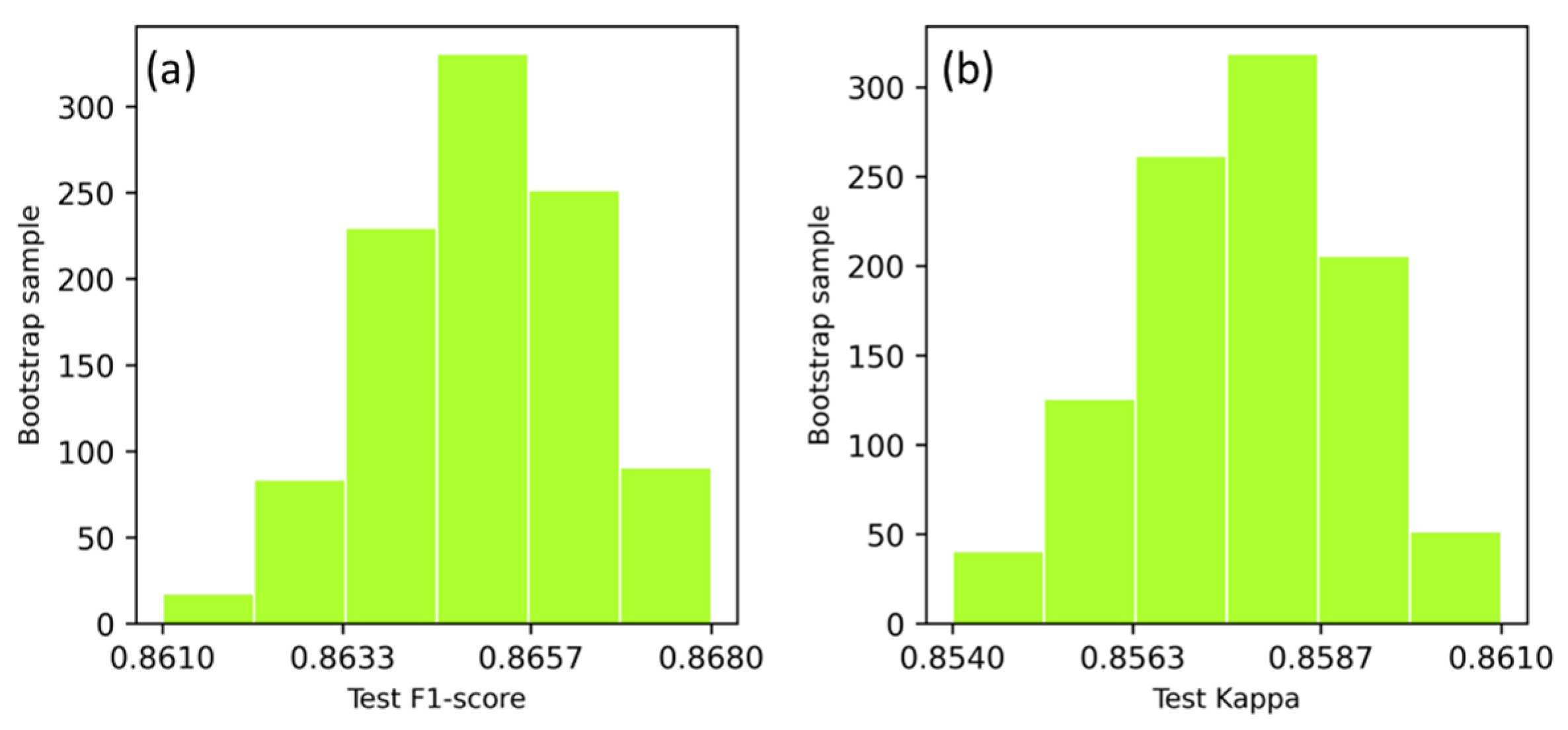
3.2. Performance of DGPE Classification
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maarel, E. Vegetation Mapping: Vegetation Science in Need of a New Handbook. J. Veg. Sci. 1991, 2, 421–424. [Google Scholar] [CrossRef]
- Mucina, L. Classification of Vegetation: Past, Present and Future. J. Veg. Sci. 1997, 8, 751–760. [Google Scholar] [CrossRef]
- Küchler, A.W.; Zonneveld, I.S. Vegetation Mapping; Springer: Dordrecht, The Netherlands, 1988; ISBN 978-94-009-3083-4. [Google Scholar]
- Grossman, D.; Faber-Langendoen, D.; Weakley, A.; Anderson, M.; Bourgeron, P.; Crawford, R.; Goodin, K.; Landaal, S.; Metzler, K.; Patterson, K. International Classification of Ecological Communities: Terrestrial Vegetation of the United States; The Nature Conservancy: Arlington County, VA, USA, 1998. [Google Scholar]
- Gleason, H.A. The Individualistic Concept of the Plant Association. Bull. Torrey Bot. Club 1926, 53, 7. [Google Scholar] [CrossRef]
- Moravec, J. Influences of the Individualistic Concept of Vegetation on Syntaxonomy. Vegetatio 1989, 81, 29–39. [Google Scholar] [CrossRef]
- Collins, S.L.; Glenn, S.M.; Roberts, D.W. The Hierarchical Continuum Concept. J. Veg. Sci. 1993, 4, 149–156. [Google Scholar] [CrossRef]
- Clements, F.E. Plant Succession: An Analysis of the Development of Vegetation; Carnegie Institution of Washington: Washington, DC, USA, 1916. [Google Scholar]
- McIntosh, R.P. The Continuum Concept of Vegetation. Bot. Rev. 1967, 33, 130–187. [Google Scholar] [CrossRef]
- Austin, M.P. Continuum Concept, Ordination Methods, and Niche Theory. Annu. Rev. Ecol. Syst. 1985, 16, 39–61. [Google Scholar] [CrossRef]
- Mitchell, R.M.; Bakker, J.D.; Vincent, J.B.; Davies, G.M. Relative Importance of Abiotic, Biotic, and Disturbance Drivers of Plant Community Structure in the Sagebrush Steppe. Ecol. Appl. 2017, 27, 756–768. [Google Scholar] [CrossRef]
- Whittaker, R.H. Classification of Plant Communities; Springer: Dordrecht, The Netherlands, 1980; ISBN 978-94-009-9183-5. [Google Scholar]
- McIntosh, R.P. Concept and Terminology of Homogeneity and Heterogeneity in Ecology. In Ecological Heterogeneity; Kolasa, J., Pickett, S.T.A., Eds.; Ecological Studies; Springer: New York, NY, USA, 1991; Volume 86, pp. 24–46. ISBN 978-1-4612-7781-1. [Google Scholar]
- Bedward, M.; Keith, D.A.; Pressey, R.L. Homogeneity Analysis: Assessing the Utility of Classifications and Maps of Natural Resources. Austral. Ecol. 1992, 17, 133–139. [Google Scholar] [CrossRef]
- Feagin, R.A. Heterogeneity versus Homogeneity: A Conceptual and Mathematical Theory in Terms of Scale-Invariant and Scale-Covariant Distributions. Ecol. Complex. 2005, 2, 339–356. [Google Scholar] [CrossRef]
- Köppen, W. Das Geographische System der Klimate. Available online: http://koeppen-geiger.vu-wien.ac.at/pdf/Koppen_1936.pdf (accessed on 20 March 2022).
- Bailey, R.G. Ecosystem Geography: From Ecoregions to Sites, 2nd ed.; Springer: New York, NY, USA, 2009; ISBN 978-0-387-89515-4. [Google Scholar]
- Metzger, M.J.; Bunce, R.G.H.; Jongman, R.H.G.; Sayre, R.; Trabucco, A.; Zomer, R. A High-Resolution Bioclimate Map of the World: A Unifying Framework for Global Biodiversity Research and Monitoring. Glob. Ecol. Biogeogr. 2013, 22, 630–638. [Google Scholar] [CrossRef]
- Küchler, A.W. A Physiognomic Classification of Vegetation. Ann. Assoc. Am. Geogr. 1949, 39, 201–210. [Google Scholar] [CrossRef]
- Beard, J.S. The Physiognomic Approach. In Classification of Plant Communities; Whittaker, R.H., Ed.; Springer: Dordrecht, The Netherlands, 1978; pp. 33–64. ISBN 978-94-009-9183-5. [Google Scholar]
- Braun-Blanquet, J. Pflanzensoziologie; Springer: Vienna, Austria, 1964; ISBN 978-3-7091-8111-9. [Google Scholar]
- Westhoff, V.; Van Der Maarel, E. The Braun-Blanquet Approach. In Classification of Plant Communities; Springer: New York, NY, USA, 1978; pp. 287–399. [Google Scholar]
- Gaston, K.J. Valuing Common Species. Science 2010, 327, 154–155. [Google Scholar] [CrossRef]
- Sharma, R.C. Genus-Physiognomy-Ecosystem (GPE) System for Satellite-Based Classification of Plant Communities. Ecologies 2021, 2, 203–213. [Google Scholar] [CrossRef]
- Sharma, R.C. Countrywide Mapping of Plant Ecological Communities with 101 Legends Including Land Cover Types for the First Time at 10 m Resolution through Convolutional Learning of Satellite Images. Appl. Sci. 2022, 12, 7125. [Google Scholar] [CrossRef]
- Sharma, R.C.; Hirayama, H.; Yasuda, M.; Asai, M.; Hara, K. Classification and Mapping of Plant Communities Using Multi-Temporal and Multi-Spectral Satellite Images. J. Geogr. Geol. 2022, 14, 43. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Mayer, B.; Kylling, A. Technical Note: The LibRadtran Software Package for Radiative Transfer Calculations—Description and Examples of Use. Atmos. Chem. Phys. 2005, 5, 1855–1877. [Google Scholar] [CrossRef]
- Richter, R.; Schläpfer, D. Atmospheric/Topographic Correction for Satellite Imagery. Available online: https://www.rese.ch/pdf/atcor3_manual.pdf (accessed on 20 March 2022).
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Dixon, P.M. Bootstrap Resampling. In Encyclopedia of Environmetrics; El-Shaarawi, A.H., Piegorsch, W.W., Eds.; Wiley: Hoboken, NJ, USA, 2001; ISBN 978-0-471-89997-6. [Google Scholar]
- Aggemyr, E.; Cousins, S.A.O. Landscape Structure and Land Use History Influence Changes in Island Plant Composition after 100 Years: Revisiting 27 Islands after 100 Years. J. Biogeogr. 2012, 39, 1645–1656. [Google Scholar] [CrossRef]
- Franklin, J.; Serra-Diaz, J.M.; Syphard, A.D.; Regan, H.M. Global Change and Terrestrial Plant Community Dynamics. Proc. Natl. Acad. Sci. USA 2016, 113, 3725–3734. [Google Scholar] [CrossRef]
- Chen, C.; Wu, S.; Meurk, C.D.; Ma, M.; Zhao, J.; Mingquan, L.; Tong, X. Effects of Local and Landscape Factors on Exotic Vegetation in the Riparian Zone of a Regulated River: Implications for Reservoir Conservation. Landsc. Urban Plan. 2017, 157, 45–55. [Google Scholar] [CrossRef]
- Prokopová, M.; Salvati, L.; Egidi, G.; Cudlín, O.; Včeláková, R.; Plch, R.; Cudlín, P. Envisioning Present and Future Land-Use Change under Varying Ecological Regimes and Their Influence on Landscape Stability. Sustainability 2019, 11, 4654. [Google Scholar] [CrossRef]
- Afuye, G.A.; Kalumba, A.M.; Busayo, E.T.; Orimoloye, I.R. A Bibliometric Review of Vegetation Response to Climate Change. Env. Sci. Pollut. Res. 2022, 29, 18578–18590. [Google Scholar] [CrossRef]
- Zeng, N.; Neelin, J.D. The Role of Vegetation–Climate Interaction and Interannual Variability in Shaping the African Savanna. J. Clim. 2000, 13, 2665–2670. [Google Scholar] [CrossRef]
- Götzenberger, L.; de Bello, F.; Bråthen, K.A.; Davison, J.; Dubuis, A.; Guisan, A.; Lepš, J.; Lindborg, R.; Moora, M.; Pärtel, M.; et al. Ecological Assembly Rules in Plant Communities-Approaches, Patterns and Prospects. Biol. Rev. 2012, 87, 111–127. [Google Scholar] [CrossRef]
- Cannone, N.; Pignatti, S. Ecological Responses of Plant Species and Communities to Climate Warming: Upward Shift or Range Filling Processes? Clim. Chang. 2014, 123, 201–214. [Google Scholar] [CrossRef]
- Diao, C.; Liu, Y.; Zhao, L.; Zhuo, G.; Zhang, Y. Regional-Scale Vegetation-Climate Interactions on the Qinghai-Tibet Plateau. Ecol. Inform. 2021, 65, 101413. [Google Scholar] [CrossRef]
- Gould, W. Remote sensing of vegetation, plant species richness, and regional biodiversity hotspots. Ecol. Appl. 2000, 10, 1861–1870. [Google Scholar] [CrossRef]
- Mathieu, R.; Aryal, J.; Chong, A. Object-Based Classification of Ikonos Imagery for Mapping Large-Scale Vegetation Communities in Urban Areas. Sensors 2007, 7, 2860–2880. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Sha, Z.; Yu, M. Remote Sensing Imagery in Vegetation Mapping: A Review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- He, K.S.; Rocchini, D.; Neteler, M.; Nagendra, H. Benefits of Hyperspectral Remote Sensing for Tracking Plant Invasions: Plant Invasion and Hyperspectral Remote Sensing. Divers. Distrib. 2011, 17, 381–392. [Google Scholar] [CrossRef]
- Watt, A.S. Pattern and Process in the Plant Community. J. Ecol. 1947, 35, 1. [Google Scholar] [CrossRef]
- Cullum, C.; Rogers, K.H.; Brierley, G.; Witkowski, E.T.F. Ecological Classification and Mapping for Landscape Management and Science: Foundations for the Description of Patterns and Processes. Prog. Phys. Geogr. Earth Environ. 2016, 40, 38–65. [Google Scholar] [CrossRef]
- Avolio, M.L.; Forrestel, E.J.; Chang, C.C.; La Pierre, K.J.; Burghardt, K.T.; Smith, M.D. Demystifying Dominant Species. New Phytol. 2019, 223, 1106–1126. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Knapp, A.K. Dominant Species Maintain Ecosystem Function with Non-Random Species Loss. Ecol. Lett. 2003, 6, 509–517. [Google Scholar] [CrossRef]
- Winfree, R.; Fox, J.W.; Williams, N.M.; Reilly, J.R.; Cariveau, D.P. Abundance of Common Species, Not Species Richness, Drives Delivery of a Real-World Ecosystem Service. Ecol. Lett. 2015, 18, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Perino, A.; Pereira, H.M.; Felipe-Lucia, M.; Kim, H.; Kühl, H.S.; Marselle, M.R.; Meya, J.N.; Meyer, C.; Navarro, L.M.; van Klink, R.; et al. Biodiversity Post-2020: Closing the Gap between Global Targets and National-level Implementation. Conserv. Lett. 2022, 15, e12848. [Google Scholar] [CrossRef]
- Hengl, T.; Walsh, M.G.; Sanderman, J.; Wheeler, I.; Harrison, S.P.; Prentice, I.C. Global Mapping of Potential Natural Vegetation: An Assessment of Machine Learning Algorithms for Estimating Land Potential. PeerJ 2018, 6, e5457. [Google Scholar] [CrossRef]
- Sato, H.; Ise, T. Predicting Global Terrestrial Biomes with the LeNet Convolutional Neural Network. Geosci. Model Dev. 2022, 15, 3121–3132. [Google Scholar] [CrossRef]
- Ellenberg, H.; Mueller-Dombois, D. Tentative Physiognomic-Ecological Classification of Plant Formations of the Earth. Available online: https://doi.org/10.5169/SEALS-377650 (accessed on 10 March 2022).
- Noriyuki, M.; Kondo, H.; Shitara, T.; Yoshikawa, M.; Hoshino, Y. A New Formal Classification for Japanese Forest Vegetation Based on Traditional Phytosociological Concepts. Appl. Veg. Sci. 2021, 24, e12611. [Google Scholar] [CrossRef]
- Sharma, R.C. An Ultra-Resolution Features Extraction Suite for Community-Level Vegetation Differentiation and Mapping at a Sub-Meter Resolution. Remote Sens. 2022, 14, 3145. [Google Scholar] [CrossRef]
- Özesmi, S.L.; Tan, C.O.; Özesmi, U. Methodological Issues in Building, Training, and Testing Artificial Neural Networks in Ecological Applications. Ecol. Model. 2006, 195, 83–93. [Google Scholar] [CrossRef]
- Brodrick, P.G.; Davies, A.B.; Asner, G.P. Uncovering Ecological Patterns with Convolutional Neural Networks. Trends Ecol. Evol. 2019, 34, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Christin, S.; Hervet, É.; Lecomte, N. Applications for Deep Learning in Ecology. Methods Ecol. Evol. 2019, 10, 1632–1644. [Google Scholar] [CrossRef]
- Ryo, M.; Angelov, B.; Mammola, S.; Kass, J.M.; Benito, B.M.; Hartig, F. Explainable Artificial Intelligence Enhances the Ecological Interpretability of Black-box Species Distribution Models. Ecography 2021, 44, 199–205. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Carpenter, G.A.; Gopal, S.; Macomber, S.; Martens, S.; Woodcock, C.E.; Franklin, J. A Neural Network Method for Efficient Vegetation Mapping. Remote Sens. Environ. 1999, 70, 326–338. [Google Scholar] [CrossRef]
- Campos-Taberner, M.; García-Haro, F.J.; Martínez, B.; Izquierdo-Verdiguier, E.; Atzberger, C.; Camps-Valls, G.; Gilabert, M.A. Understanding Deep Learning in Land Use Classification Based on Sentinel-2 Time Series. Sci. Rep. 2020, 10, 17188. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Z.; Zhang, X.; Liu, Y.; Sun, X.; Pang, C.; Zhao, C. Vegetation Land Use/Land Cover Extraction from High-Resolution Satellite Images Based on Adaptive Context Inference. IEEE Access 2020, 8, 21036–21051. [Google Scholar] [CrossRef]
- Bakhti, K.; El Amin Larabi, M. Comparing Deep Recurrent Learning and Convolutional Learning for Multi-Temporal Vegetation Classification. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 4392–4395. [Google Scholar]
- Neves, A.K.; Körting, T.S.; Fonseca, L.M.G.; Soares, A.R.; Girolamo-Neto, C.D.; Heipke, C. Hierarchical Mapping of Brazilian Savanna (Cerrado) Physiognomies Based on Deep Learning. J. Appl. Rem. Sens. 2021, 15, 044504. [Google Scholar] [CrossRef]
- Ienco, D.; Gaetano, R.; Dupaquier, C.; Maurel, P. Land Cover Classification via Multitemporal Spatial Data by Deep Recurrent Neural Networks. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1685–1689. [Google Scholar] [CrossRef]
- Lakhal, M.I.; Çevikalp, H.; Escalera, S.; Ofli, F. Recurrent Neural Networks for Remote Sensing Image Classification. IET Comput. Vis. 2018, 12, 1040–1045. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, X.; Zhang, X.; Wu, D.; Du, X. Long Time Series Land Cover Classification in China from 1982 to 2015 Based on Bi-LSTM Deep Learning. Remote Sens. 2019, 11, 1639. [Google Scholar] [CrossRef]
- Sun, Z.; Di, L.; Fang, H. Using Long Short-Term Memory Recurrent Neural Network in Land Cover Classification on Landsat and Cropland Data Layer Time Series. Int. J. Remote Sens. 2019, 40, 593–614. [Google Scholar] [CrossRef]
- Mishra, A.P.; Rai, I.D.; Pangtey, D.; Padalia, H. Vegetation Characterization at Community Level Using Sentinel-2 Satellite Data and Random Forest Classifier in Western Himalayan Foothills, Uttarakhand. J. Indian Soc. Remote Sens. 2021, 49, 759–771. [Google Scholar] [CrossRef]
- Adagbasa, E.G.; Adelabu, S.A.; Okello, T.W. Application of Deep Learning with Stratified K-Fold for Vegetation Species Discrimation in a Protected Mountainous Region Using Sentinel-2 Image. Geocarto Int. 2022, 37, 142–162. [Google Scholar] [CrossRef]
- Kluczek, M.; Zagajewski, B.; Kycko, M. Airborne HySpex Hyperspectral Versus Multitemporal Sentinel-2 Images for Mountain Plant Communities Mapping. Remote Sens. 2022, 14, 1209. [Google Scholar] [CrossRef]
- Martínez Prentice, R.; Villoslada Peciña, M.; Ward, R.D.; Bergamo, T.F.; Joyce, C.B.; Sepp, K. Machine Learning Classification and Accuracy Assessment from High-Resolution Images of Coastal Wetlands. Remote Sens. 2021, 13, 3669. [Google Scholar] [CrossRef]

| DSPE | Inference | DGPE |
|---|---|---|
| Species-Physiognomy | Abies Ecf |
| Species-Physiognomy | Abies Ecf |
| Species-Physiognomy | Abies Ecf |
| Species-Physiognomy | Acer Dbf |
| Species-Physiognomy | Acer Sb |
| Species-Physiognomy | Alnus Dbf |
| Species-Physiognomy | Alnus Dsb |
| Physiognomy-Ecological | Alpine Hb |
| Physiognomy-Ecological | Alpine Sb |
| Physiognomy-Ecological | Bamboo Ebf |
| Species-Physiognomy | Betula Dbf |
| Species-Physiognomy | Betula Dbf |
| Species-Physiognomy | Betula Dbf |
| Species-Physiognomy (Multi strata) | Betula Dbf |
| Species-Physiognomy | Camellia Ebf |
| Species-Physiognomy | Carpinus Dbf |
| Species-Physiognomy | Castanopsis Ebf |
| Physiognomy-Ecological | Coastal Hb |
| Physiognomy-Ecological | Coastal Sb |
| Species-Physiognomy | Cryptomeria Ecf |
| Species-Physiognomy (Multi strata) | Fagus Dbf |
| Species-Physiognomy | Euptelea Dbf |
| Species-Physiognomy | Fagus Dbf |
| Species-Physiognomy | Fagus Dbf |
| Species-Physiognomy | Fraxinus Dbf |
| Species-Physiognomy | Hydrangea Sb |
| Species-Physiognomy | Juglans Dbf |
| Species-Physiognomy | Larix Dcf |
| Species-Physiognomy | Machilus Ebf |
| Species-Physiognomy | Mallotus Dbf |
| Species-Physiognomy | Miscanthus Hb |
| Physiognomy-Ecological | Open-space Hb |
| Species-Physiognomy (Multi strata) | Quercus Dbf |
| Species-Physiognomy | Picea Ecf |
| Species-Physiognomy | Pinus Ecf |
| Species-Physiognomy | Pinus Ecf |
| Species-Physiognomy | Pinus Sb |
| Species-Physiognomy | Pinus Ecf |
| Species-Physiognomy | Populus Dbf |
| Species-Physiognomy | Pterocarya Dbf |
| Species-Physiognomy | Quercus Dbf |
| Species-Physiognomy | Quercus Dbf |
| Species-Physiognomy | Quercus Ebf |
| Species-Physiognomy | Quercus Dbf |
| Species-Physiognomy | Quercus Sb |
| Species-Physiognomy | Rhododendron Sb |
| Species-Physiognomy | Robinia Dbf |
| Species-Physiognomy | Salix Dbf |
| Species-Physiognomy | Salix Sb |
| Species-Physiognomy | Sasa Sb |
| Species-Physiognomy | Thuja Ecf |
| Species-Physiognomy | Thujopsis Ecf |
| Species-Physiognomy | Tilia Dbf |
| Species-Physiognomy | Tsuga Ecf |
| Species-Physiognomy | Ulmus Dbf |
| Species-Physiognomy | Weigela Sb |
| Physiognomy-Ecological | Wetland Hb |
| Species-Physiognomy | Zelkova Dbf |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, R.C. Dominant Species-Physiognomy-Ecological (DSPE) System for the Classification of Plant Ecological Communities from Remote Sensing Images. Ecologies 2022, 3, 323-335. https://doi.org/10.3390/ecologies3030025
Sharma RC. Dominant Species-Physiognomy-Ecological (DSPE) System for the Classification of Plant Ecological Communities from Remote Sensing Images. Ecologies. 2022; 3(3):323-335. https://doi.org/10.3390/ecologies3030025
Chicago/Turabian StyleSharma, Ram C. 2022. "Dominant Species-Physiognomy-Ecological (DSPE) System for the Classification of Plant Ecological Communities from Remote Sensing Images" Ecologies 3, no. 3: 323-335. https://doi.org/10.3390/ecologies3030025






