Ozonized Oleic Acid as a New Viticultural Treatment? Study of the Effect of LIQUENSO® Oxygenate on the Carpoplane Microbial Community and Wine Microorganisms Combining Metabarcoding and In Vitro Assays
Abstract
:1. Introduction
2. Experimental Section
2.1. Ozonides
2.2. Agrochemical Treatments
2.3. Wine Grape Sampling
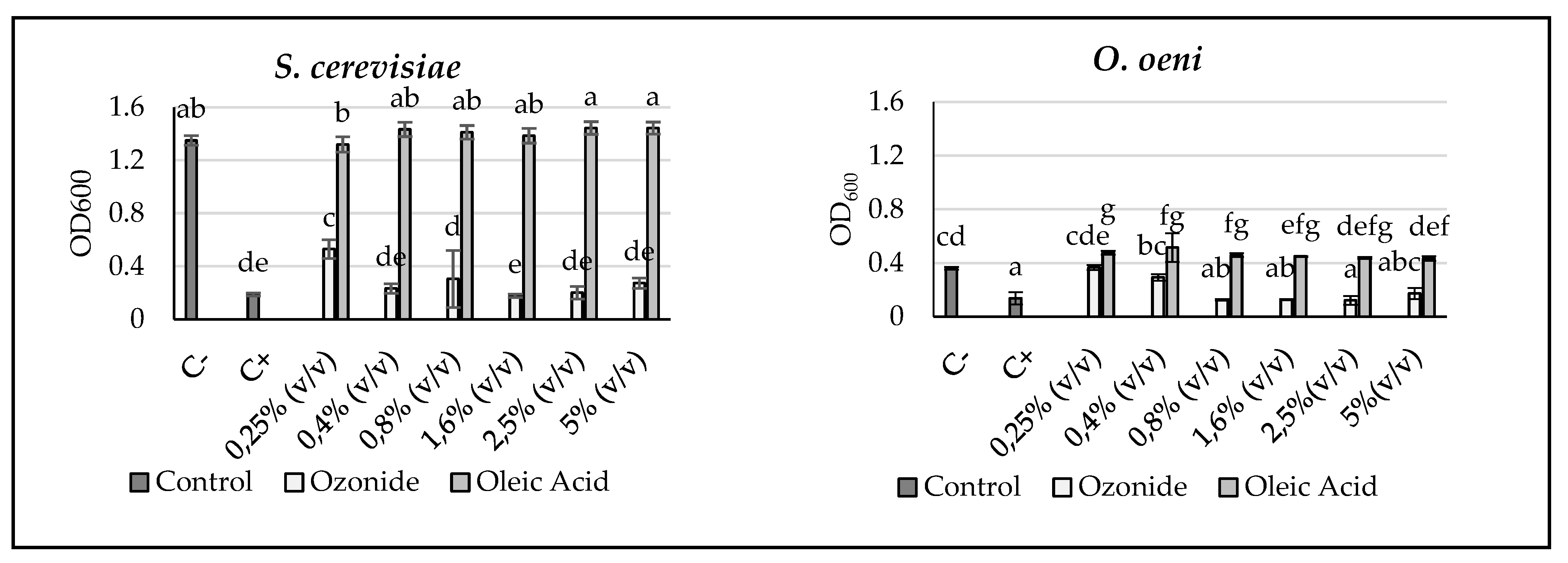
2.4. 96-Well Microtiter Assays
2.5. DNA Extraction and NGS Sequencing
2.6. Analysis of the NGS Data
3. Results
3.1. Microecosystem Effects of Ozonized Oleic Acid Treatment in the Vineyard
3.2. Community Structure
3.3. Efficiency Analyses against Yeasts and Bacteria Relevant to the Vinification Process
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The Microbial Ecology of Wine Grape Berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Cavalieri, D. Metagenomic Approaches to Investigate the Contribution of the Vineyard Environment to the Quality of Wine Fermentation: Potentials and Difficulties. Front. Microbiol. 2018, 9, 991. [Google Scholar] [CrossRef]
- Escribano-Viana, R.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A.R.; González-Arenzana, L. Impact of Chemical and Biological Fungicides Applied to Grapevine on Grape Biofilm, Must, and Wine Microbial Diversity. Front. Microbiol. 2018, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Dapporto, L.; Legras, J.-L.; Calabretta, A.; Paola, M.D.; Filippo, C.D.; Viola, R.; Capretti, P.; Polsinelli, M.; Turillazzi, S.; et al. Role of Social Wasps in Saccharomyces Cerevisiae Ecology and Evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 13398–13403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Zhang, P.; Chen, D.; Howell, K. From the Vineyard to the Winery: How Microbial Ecology Drives Regional Distinctiveness of Wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef] [PubMed]
- Vitulo, N.; Lemos, W.J.F.; Calgaro, M.; Confalone, M.; Felis, G.E.; Zapparoli, G.; Nardi, T. Bark and Grape Microbiome of Vitis Vinifera: Influence of Geographic Patterns and Agronomic Management on Bacterial Diversity. Front. Microbiol. 2019, 9, 3203. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-J.; Delmotte, F.; Cervera, S.R.; Douence, L.; Greif, C.; Corio-Costet, M.-F. At Least Two Origins of Fungicide Resistance in Grapevine Downy Mildew Populations. Appl. Environ. Microbiol. 2007, 73, 5162–5172. [Google Scholar] [CrossRef] [Green Version]
- Leroch, M.; Kretschmer, M.; Hahn, M. Fungicide Resistance Phenotypes of Botrytis Cinerea Isolates from Commercial Vineyards in South West Germany. J. Phytopathol. 2011, 159, 63–65. [Google Scholar] [CrossRef]
- Delmas, C.E.L.; Dussert, Y.; Delière, L.; Couture, C.; Mazet, I.D.; Richart Cervera, S.; Delmotte, F. Soft Selective Sweeps in Fungicide Resistance Evolution: Recurrent Mutations without Fitness Costs in Grapevine Downy Mildew. Mol. Ecol. 2017, 26, 1936–1951. [Google Scholar] [CrossRef]
- de Almeida Kogawa, N.R.; de Arruda, E.J.; Micheletti, A.C.; Matos, M.D.F.C.; de Oliveira, L.C.S.; de Lima, D.P.; Carvalho, N.C.P.; de Oliveira, P.D.; de Castro Cunha, M.; Ojeda, M. Synthesis, Characterization, Thermal Behavior, and Biological Activity of Ozonides from Vegetable Oils. RSC Adv. 2015, 5, 65427–65436. [Google Scholar] [CrossRef]
- Günaydın, Y.; Sevim, H.; Tanyolaç, D.; Gürpınar, Ö.A. Ozonated Olive Oil with a High Peroxide Value for Topical Applications: In-Vitro Cytotoxicity Analysis with L929 Cells. Ozone Sci. Eng. 2018, 40, 37–43. [Google Scholar] [CrossRef]
- Sechi, L.A.; Lezcano, I.; Nunez, N.; Espim, M.; Duprè, I.; Pinna, A.; Molicotti, P.; Fadda, G.; Zanetti, S. Antibacterial Activity of Ozonized Sunflower Oil (Oleozon). J. Appl. Microbiol. 2001, 90, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Geweely, N.S.I. Antifungal Activity of Ozonized Olive Oil (Oleozone). Int. J. Agri. Biol. 2006, 8, 671–678. [Google Scholar]
- Ma, Y.; Ke, Y.; Zhu, H.; Li, B.; Li, B.; Zhang, F.; Li, Y. Oleozon: A Novel Control Strategy against Powdery Mildew in Cucumber. J. Phytopathol. 2017, 165, 841–847. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Mancabelli, L.; Milani, C.; Ventura, M.; Ciani, M.; Comitini, F. The Influence of Fungicide Treatments on Mycobiota of Grapes and Its Evolution during Fermentation Evaluated by Metagenomic and Culture-Dependent Methods. Microorganisms 2019, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, J.A.; van der Lelie, D.; Zarraonaindia, I. Microbial Terroir for Wine Grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef] [Green Version]
- Capozzi, V.; Tufariello, M.; De Simone, N.; Fragasso, M.; Grieco, F. Biodiversity of Oenological Lactic Acid Bacteria: Species-and Strain-Dependent plus/Minus Effects on Wine Quality and Safety. Fermentation 2021, 7, 24. [Google Scholar] [CrossRef]
- Strickland, M.T.; Schopp, L.M.; Edwards, C.G.; Osborne, J.P. Impact of Pediococcus spp. on Pinot Noir Wine Quality and Growth of Brettanomyces. Am. J. Enol. Vitic. 2016, 67, 188–198. [Google Scholar] [CrossRef]
- Wade, M.E.; Strickland, M.T.; Osborne, J.P.; Edwards, C.G. Role of Pediococcus in Winemaking. Aust. J. Grape Wine Res. 2019, 25, 7–24. [Google Scholar] [CrossRef] [Green Version]
- Impact of Different Temperature Profiles on Simultaneous Yeast and Bacteria Fermentation|SpringerLink. Available online: https://link.springer.com/article/10.1186/s13213-020-01565-w (accessed on 23 June 2022).
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Growth of Non-Saccharomyces Yeasts Affects Nutrient Availability for Saccharomyces Cerevisiae during Wine Fermentation. Int. J. Food Microbiol. 2012, 157, 245–250. [Google Scholar] [CrossRef]
- Vicente Sánchez, J.; Calderón, F.; Santos de la Sen, A.; Marquina Díaz, D.; Benito, S. High Potential of Pichia Kluyveri and Other Pichia Species in Wine Technology. Int. J. Mol. Sci. 2021, 22, 1196. [Google Scholar] [CrossRef] [PubMed]
- Snowdon, E.M.; Bowyer, M.C.; Grbin, P.R.; Bowyer, P.K. Mousy Off-Flavor: A Review. J. Agric. Food Chem. 2006, 54, 6465–6474. [Google Scholar] [CrossRef] [PubMed]
- Costello, P.J.; Siebert, T.E.; Solomon, M.R.; Bartowsky, E.J. Synthesis of Fruity Ethyl Esters by Acyl Coenzyme A: Alcohol Acyltransferase and Reverse Esterase Activities in Oenococcus Oeni and Lactobacillus Plantarum. J. Appl. Microbiol. 2013, 114, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Morata, A.; Loira, I.; González, C.; Escott, C. Non-Saccharomyces as Biotools to Control the Production of Off-Flavors in Wines. Molecules 2021, 26, 4571. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected Non-Saccharomyces Wine Yeasts in Controlled Multistarter Fermentations with Saccharomyces Cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef]
- Costello, P.J.; Lee, T.H.; Henschke, P. Ability of Lactic Acid Bacteria to Produce N-Heterocycles Causing Mousy off-Flavour in Wine. Aust. J. Grape Wine Res. 2001, 7, 160–167. [Google Scholar] [CrossRef]
- Romano, A.; Perello, M.C.; de Revel, G.; Lonvaud-Funel, A. Growth and Volatile Compound Production by Brettanomyces/Dekkera Bruxellensis in Red Wine. J. Appl. Microbiol. 2008, 104, 1577–1585. [Google Scholar] [CrossRef]
- Virdis, C.; Sumby, K.; Bartowsky, E.; Jiranek, V. Lactic Acid Bacteria in Wine: Technological Advances and Evaluation of Their Functional Role. Front. Microbiol. 2021, 11, 612118. [Google Scholar] [CrossRef]
- Pérez-Martín, F.; Seseña, S.; Izquierdo, P.M.; Martín, R.; Palop, M.L. Screening for Glycosidase Activities of Lactic Acid Bacteria as a Biotechnological Tool in Oenology. World J. Microbiol. Biotechnol. 2012, 28, 1423–1432. [Google Scholar] [CrossRef]
- Benito, S. The Management of Compounds That Influence Human Health in Modern Winemaking from an HACCP Point of View. Fermentation 2019, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Mamlouk, D.; Gullo, M. Acetic Acid Bacteria: Physiology and Carbon Sources Oxidation. Indian J. Microbiol. 2013, 53, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Lynch, K.M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E.K. Physiology of Acetic Acid Bacteria and Their Role in Vinegar and Fermented Beverages. Compr. Rev. Food Sci. Food Saf. 2019, 18, 587–625. [Google Scholar] [CrossRef] [Green Version]
- Shinjoh, M.; Toyama, H. Industrial Application of Acetic Acid Bacteria (Vitamin C and Others). In Acetic Acid Bacteria: Ecology and Physiology; Matsushita, K., Toyama, H., Tonouchi, N., Okamoto-Kainuma, A., Eds.; Springer: Japan, Tokyo, 2016; pp. 321–338. ISBN 978-4-431-55933-7. [Google Scholar]
- Liu, L.; Chen, Y.; Yu, S.; Chen, J.; Zhou, J. Simultaneous Transformation of Five Vectors in Gluconobacter Oxydans. Plasmid 2021, 117, 102588. [Google Scholar] [CrossRef]
- Soares-Santos, V.; Pardo, I.; Ferrer, S. Cells-QPCR as a Direct Quantitative PCR Method to Avoid Microbial DNA Extractions in Grape Musts and Wines. Int. J. Food Microbiol. 2017, 261, 25–34. [Google Scholar] [CrossRef]
- Brysch-Herzberg, M.; Seidel, M. Yeast Diversity on Grapes in Two German Wine Growing Regions. Int. J. Food Microbiol. 2015, 214, 137–144. [Google Scholar] [CrossRef]
- Thijs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative Evaluation of Four Bacteria-Specific Primer Pairs for 16S RRNA Gene Surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-Coverage ITS Primers for the DNA-Based Identification of Ascomycetes and Basidiomycetes in Environmental Samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [Green Version]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the Diversity of Grapevine Microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef] [Green Version]
- Petit, B.; Mitaine-Offer, A.-C.; Fischer, J.; Schüffler, A.; Delaude, C.; Miyamoto, T.; Tanaka, C.; Thines, E.; Lacaille-Dubois, M.-A. Anti-Phytopathogen Terpenoid Glycosides from the Root Bark of Chytranthus Macrobotrys and Radlkofera Calodendron. Phytochemistry 2021, 188, 112797. [Google Scholar] [CrossRef]
- Xu, Y.; Xie, M.; Xue, J.; Xiang, L.; Li, Y.; Xiao, J.; Xiao, G.; Wang, H.-L. EGCG Ameliorates Neuronal and Behavioral Defects by Remodeling Gut Microbiota and TotM Expression in Drosophila Models of Parkinson’s Disease. FASEB J. 2020, 34, 5931–5950. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Bagheri, B.; Morgan, H.H.; Divol, B.; Setati, M.E. Assessment of Wine Microbial Diversity Using ARISA and Cultivation-Based Methods. Ann. Microbiol. 2015, 65, 1833–1840. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finotello, F.; Mastrorilli, E.; Di Camillo, B. Measuring the Diversity of the Human Microbiota with Targeted Next-Generation Sequencing. Brief. Bioinform. 2018, 19, 679–692. [Google Scholar] [CrossRef]
- Hill, T.C.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using Ecological Diversity Measures with Bacterial Communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef]
- User’s Guide to Correlation Coefficients|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S2452247318302164?token=F352ED4A7123774FDFAC340E8319705D27C1CD7A79AC08AE39A025621711EC86BFED8F2367A02DB9758D6215D239487B&originRegion=eu-west-1&originCreation=20220621145200 (accessed on 21 June 2022).
- Liu, D.; Howell, K. Community Succession of the Grapevine Fungal Microbiome in the Annual Growth Cycle. Environ. Microbiol. 2021, 23, 1842–1857. [Google Scholar] [CrossRef]
- Moubasher, A.-A.H.; Abdel-Sater, M.A.; Soliman, Z. Biodiversity of Filamentous and Yeast Fungi in Citrus and Grape Fruits and Juices in Assiut Area, Egypt. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 353–365. [Google Scholar] [CrossRef]
- Garijo, P.; López, R.; Santamaría, P.; Ocón, E.; Olarte, C.; Sanz, S.; Gutiérrez, A.R. Presence of Enological Microorganisms in the Grapes and the Air of a Vineyard during the Ripening Period. Eur. Food Res. Technol. 2011, 233, 359–365. [Google Scholar] [CrossRef]
- Renouf, V.; Claisse, O.; Lonvaud-Funel, A. Understanding the Microbial Ecosystem on the Grape Berry Surface through Numeration and Identification of Yeast and Bacteria. Aust. J. Grape Wine Res. 2005, 11, 316–327. [Google Scholar] [CrossRef]
- Martins, G.; Vallance, J.; Mercier, A.; Albertin, W.; Stamatopoulos, P.; Rey, P.; Lonvaud, A.; Masneuf-Pomarède, I. Influence of the Farming System on the Epiphytic Yeasts and Yeast-like Fungi Colonizing Grape Berries during the Ripening Process. Int. J. Food Microbiol. 2014, 177, 21–28. [Google Scholar] [CrossRef]
- Ding, Y.; Wei, R.; Wang, L.; Yang, C.; Li, H.; Wang, H. Diversity and Dynamics of Microbial Ecosystem on Berry Surface during the Ripening of Ecolly (Vitis Vinifera L.) Grape in Wuhai, China. World J. Microbiol. Biotechnol. 2021, 37, 214. [Google Scholar] [CrossRef]
- Abdullabekova, D.A.; Magomedova, E.S.; Aliverdieva, D.A.; Kachalkin, A.V. Yeast Communities of Vineyards in Dagestan: Ecological, Taxonomic, and Genetic Characteristics. Biol. Bull. 2020, 47, 344–351. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial Biogeography of Wine Grapes Is Conditioned by Cultivar, Vintage, and Climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Chen, Q.; Zhang, P.; Chen, D.; Howell, K.S. The Fungal Microbiome Is an Important Component of Vineyard Ecosystems and Correlates with Regional Distinctiveness of Wine. mSphere 2020, 5, e00534-20. [Google Scholar] [CrossRef]
- Boiu-Sicuia, O.-A.; Dinu, S.; Barbu, L. Physiological Profile of Some Pathogenic Bacteria Associated with Grapevine Crown Gall. Sci. Pap.-Ser. B Hortic. 2020, 64, 230–237. [Google Scholar]
- Jiang, L.; Jeong, J.C.; Lee, J.-S.; Park, J.M.; Yang, J.-W.; Lee, M.H.; Choi, S.H.; Kim, C.Y.; Kim, D.-H.; Kim, S.W.; et al. Potential of Pantoea Dispersa as an Effective Biocontrol Agent for Black Rot in Sweet Potato. Sci. Rep. 2019, 9, 16354. [Google Scholar] [CrossRef] [Green Version]
- Smits, T.H.M.; Duffy, B.; Blom, J.; Ishimaru, C.A.; Stockwell, V.O. Pantocin A, a Peptide-Derived Antibiotic Involved in Biological Control by Plant-Associated Pantoea Species. Arch. Microbiol. 2019, 201, 713–722. [Google Scholar] [CrossRef]
- Magnin-Robert, M.; Quantinet, D.; Couderchet, M.; Aziz, A.; Trotel-Aziz, P. Differential Induction of Grapevine Resistance and Defense Reactions against Botrytis Cinerea by Bacterial Mixtures in Vineyards. BioControl 2013, 58, 117–131. [Google Scholar] [CrossRef]
- Gasser, F.; Schildberger, B.; Berg, G. Biocontrol of Botrytis Cinerea by Successful Introduction of Pantoea Ananatis in the Grapevine Phyllosphere. Int. J. Wine Res. 2012, 4, 53. [Google Scholar] [CrossRef] [Green Version]
- Diversity of Bacterial Endophytes in 3 and 15 Year-Old Grapevines of Vitis Vinifera Cv. Corvina and Their Potential for Plant Growth Promotion and Phytopathogen Control|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S0944501315300318?token=6FE35BF072795D0CCD3B247C99B924899D2DE0E2FA11E3C63D920B060ADE84803E9A4DA8A6ED42BB33856BD929AC76FE&originRegion=eu-west-1&originCreation=20220620121031 (accessed on 20 June 2022).
- Whitelaw-Weckert, M.A.; Whitelaw, E.S.; Rogiers, S.Y.; Quirk, L.; Clark, A.C.; Huang, C.X. Bacterial Inflorescence Rot of Grapevine Caused by Pseudomonas Syringae Pv. Syringae. Plant Pathol. 2011, 60, 325–337. [Google Scholar] [CrossRef]
- Lipps, S.M.; Samac, D.A. Pseudomonas Viridiflava: An Internal Outsider of the Pseudomonas Syringae Species Complex. Mol. Plant Pathol. 2022, 23, 3–15. [Google Scholar] [CrossRef]
- Gerin, D.; Cariddi, C.; de Miccolis Angelini, R.M.; Rotolo, C.; Dongiovanni, C.; Faretra, F.; Pollastro, S. First Report of Pseudomonas Grapevine Bunch Rot Caused by Pseudomonas Syringae Pv. Syringae. Plant Dis. 2019, 103, 1954–1960. [Google Scholar] [CrossRef]
- Hall, S.J.; Dry, I.B.; Gopurenko, D.; Whitelaw-Weckert, M.A. Pseudomonas Syringae Pv. Syringae from Cool Climate Australian Grapevine Vineyards: New Phylogroup PG02f Associated with Bacterial Inflorescence Rot. Plant Pathol. 2019, 68, 312–322. [Google Scholar] [CrossRef]
- Marinho, M.D.C.; Diogo, B.S.; Lage, O.M.; Antunes, S.C. Ecotoxicological Evaluation of Fungicides Used in Viticulture in Non-Target Organisms. Environ. Sci. Pollut. Res. 2020, 27, 43958–43969. [Google Scholar] [CrossRef]
- Perazzolli, M.; Antonielli, L.; Storari, M.; Puopolo, G.; Pancher, M.; Giovannini, O.; Pindo, M.; Pertot, I. Resilience of the Natural Phyllosphere Microbiota of the Grapevine to Chemical and Biological Pesticides. Appl. Environ. Microbiol. 2014, 80, 3585–3596. [Google Scholar] [CrossRef] [Green Version]
- Puglisi, E.; Vasileiadis, S.; Demiris, K.; Bassi, D.; Karpouzas, D.G.; Capri, E.; Cocconcelli, P.S.; Trevisan, M. Impact of Fungicides on the Diversity and Function of Non-Target Ammonia-Oxidizing Microorganisms Residing in a Litter Soil Cover. Microb. Ecol. 2012, 64, 692–701. [Google Scholar] [CrossRef]
- Katsoula, A.; Vasileiadis, S.; Sapountzi, M.; Karpouzas, D.G. The Response of Soil and Phyllosphere Microbial Communities to Repeated Application of the Fungicide Iprodione: Accelerated Biodegradation or Toxicity? FEMS Microbiol. Ecol. 2020, 96, fiaa056. [Google Scholar] [CrossRef]
- Anand, S.K. A Comparative Analysis of Antimicrobial Property of Wine and Ozone with Calcium Hydroxide and Chlorhexidine. J. Clin. Diagn. Res. 2015, 9, ZC04-6. [Google Scholar] [CrossRef] [PubMed]
- Kadhum, A.A.H.; Wasmi, B.A.; Mohamad, A.B.; Al-Amiery, A.A.; Takriff, M.S. Preparation, Characterization, and Theoretical Studies of Azelaic Acid Derived from Oleic Acid by Use of a Novel Ozonolysis Method. Res. Chem. Intermed. 2012, 38, 659–668. [Google Scholar] [CrossRef]
- Zahardis, J.; Petrucci, G.A. The Oleic Acid-Ozone Heterogeneous Reaction System: Products, Kinetics, Secondary Chemistry, and Atmospheric Implications of a Model System—A Review. Atmos. Chem. Phys. 2007, 7, 1238–1240, 1244–1245, 1263–1265. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Available online: https://era.library.ualberta.ca/items/b4736051-ee09-49ec-8146-8f05975db42c (accessed on 8 February 2022).
- Guerrer, L.V.; Cunha, K.C.; Nogueira, M.C.L.; Cardoso, C.C.; Soares, M.M.C.N.; Almeida, M.T.G. “In Vitro” Antifungal Activity of Ozonized Sunflower Oil on Yeasts from Onychomycosis. Braz. J. Microbiol. 2012, 43, 1315–1318. [Google Scholar] [CrossRef] [Green Version]
- Monzillo, V.; Lallitto, F.; Russo, A.; Poggio, C.; Scribante, A.; Arciola, C.R.; Bertuccio, F.R.; Colombo, M. Ozonized Gel Against Four Candida Species: A Pilot Study and Clinical Perspectives. Materials 2020, 13, 1731. [Google Scholar] [CrossRef] [Green Version]
- Díaz, M.F.; Hernández, R.; Martínez, G.; Vidal, G.; Gómez, M.; Fernández, H.; Garcés, R. Comparative Study of Ozonized Olive Oil and Ozonized Sunflower Oil. J. Braz. Chem. Soc. 2006, 17, 403–407. [Google Scholar] [CrossRef]
- Torija, M.J.; Mateo, E.; Guillamón, J.M.; Mas, A. Identification and Quantification of Acetic Acid Bacteria in Wine and Vinegar by TaqMan–MGB Probes. Food Microbiol. 2010, 27, 257–265. [Google Scholar] [CrossRef]
- Kántor, A.; Kačániová, M.; Petrová, J.; Medo, J.; Hleba, L.; Rovná, K. Application of Rt-Pcr for Acetobacter Species Detection in Red Wine. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 231–234. [Google Scholar]
- Sanchez, G.M.; Fernandez, O.S.L.; Rodriguez, C. Toxicidad Aguda Dermica Del Aceite Ozonizado ‘Oleozon’ En Ratas y Conejos Rol de Los Radicales Libres CENIC. Cienc. BioloÁgicas 1997, 28, 35. [Google Scholar]
- Curtiellas, V.; Ledea, O.; Rodríguez, S.; Ancheta, O.; Echevarría, M.; Sánchez, E.; Fernández, I. El OLEOZON® Sobre La Viabilidad, La Permeabilidad Celular y La Ultraestructura de Staphylococcus Aureus. Rev. CENIC Cienc. Biol. 2008, 39, 128–131. [Google Scholar]
- Guzzon, R.; Nardin, T.; Micheletti, O.; Nicolini, G.; Larcher, R. Antimicrobial Activity of Ozone. Effectiveness against the Main Wine Spoilage Microorganisms and Evaluation of Impact on Simple Phenols in Wine. Aust. J. Grape Wine Res. 2013, 19, 180–188. [Google Scholar] [CrossRef]
- Criegee, R. Mechanismus der Ozonolyse. Angew. Chem. 1975, 87, 765–771. [Google Scholar] [CrossRef]
- Bailey, P.S. The Reactions of Ozone with Organic Compounds. Chem. Rev. 1958, 58, 925–1010. [Google Scholar] [CrossRef]
- Ziemann, P.J. Aerosol Products, Mechanisms, and Kinetics of Heterogeneous Reactions of Ozone with Oleic Acid in Pure and Mixed Particles. Faraday Discuss. 2005, 130, 469–490. [Google Scholar] [CrossRef]
- Zhou, Z.; Abbatt, J.P. Formation of Gas-Phase Hydrogen Peroxide via Multiphase Ozonolysis of Unsaturated Lipids. Environ. Sci. Technol. Lett. 2020, 8, 114–120. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhou, S.; Abbatt, J.P.D. Kinetics and Condensed-Phase Products in Multiphase Ozonolysis of an Unsaturated Triglyceride. Environ. Sci. Technol. 2019, 53, 12467–12475. [Google Scholar] [CrossRef]
- Woden, B.; Skoda, M.W.A.; Milsom, A.; Gubb, C.; Maestro, A.; Tellam, J.; Pfrang, C. Ozonolysis of Fatty Acid Monolayers at the Air–Water Interface: Organic Films May Persist at the Surface of Atmospheric Aerosols. Atmos. Chem. Phys. 2021, 21, 1325–1340. [Google Scholar] [CrossRef]
| Treatment | Richness | α-Diversity Indices | ||||
|---|---|---|---|---|---|---|
| OTU | Species | Shannon (H) | Evenness (J) | Inverse Simpson (1-D) | ||
| Portugieser I-16S | NT | 131 | 113 | 1.45 | 0.31 | 0.50 |
| CT | 127 | 118 | 1.51 | 0.32 | 0.52 | |
| OT | 50 | 43 | 0.76 | 0.20 | 0.33 | |
| Portugieser II-16S | NT | 43 | 40 | 1.98 | 0.54 | 0.81 |
| CT | 41 | 38 | 2.04 | 0.56 | 0.82 | |
| OT | 32 | 29 | 1.36 | 0.40 | 0.62 | |
| Portugieser I-ITS2 | NT | 94 | 83 | 1.73 | 0.39 | 0.76 |
| CT | 94 | 80 | 1.83 | 0.42 | 0.76 | |
| OT | 74 | 66 | 1.81 | 0.43 | 0.77 | |
| Portugieser II-ITS2 | NT | 43 | 43 | 1.90 | 0.50 | 0.77 |
| CT | 44 | 41 | 1.31 | 0.35 | 0.57 | |
| OT | 43 | 41 | 1.79 | 0.48 | 0.76 | |
| Comparison of Treatments | ß-Diversity Indices | ||
|---|---|---|---|
| Sørensen Similarity Index (β1) | Jaccard Similarity Index (β2) | ||
| Portugieser I-16S | NT/CT | 0.62 | 0.45 |
| NT/OT | 0.50 | 0.33 | |
| CT/OT | 0.47 | 0.31 | |
| Portugieser II-16S | NT/CT | 0.74 | 0.59 |
| NT/OT | 0.70 | 0.53 | |
| CT/OT | 0.72 | 0.56 | |
| Portugieser I-ITS2 | NT/CT | 0.56 | 0.39 |
| NT/OT | 0.64 | 0.48 | |
| CT/OT | 0.59 | 0.42 | |
| Portugieser II-ITS2 | NT/CT | 0.60 | 0.42 |
| NT/OT | 0.71 | 0.56 | |
| CT/OT | 0.56 | 0.39 | |
| 0.25% (v/v) Ozonide | 0.4% (v/v) Ozonide | 0.8% (v/v) Ozonide | 1.6% (v/v) Ozonide | 2.5% (v/v) Ozonide | 5% (v/v) Ozonide | C− | C+ | |
|---|---|---|---|---|---|---|---|---|
| Acetobacter aceti | 19.9 ± 6.6 | 3.6 ± 5.2 | 3.4 ± 6.5 | −0.4 ± 3.1 | 6.8 ± 5.8 | 2.6 ± 2.8 | 100.0 ± 3.2 | 0.0 ± 5.1 |
| Gluconobacter oxydans | 108.8 ± 25.6 | 49.3 ± 32.3 | 5.6 ± 3.0 | −9.7 ± 1.6 | −10.5 ± 4.5 | −7.9 ± 0.2 | 100.0 ± 17.0 | 0.0 ± 8.8 |
| Levilactobacillus brevis | 125.1 ± 13.8 | 116.2 ± 2.6 | 111.1 ± 4.5 | 56.4 ± 1.6 | 76.5 ± 3.0 | 35.4 ± 2.3 | 100.0 ± 9.1 | 0.0 ± 2.5 |
| Lactiplantibacillus plantarum | 102.3 ± 6.4 | 89.1 ± 8.3 | 80.1 ± 2.9 | 49.8 ± 4.4 | 61.0 ± 1.9 | 40.4 ± 1.7 | 100.0 ± 4.5 | 0.0 ± 1.7 |
| Oenococcus oeni | 102.1 ±7.29 | 72.4 ± 10.0 | 4.3± 2.7 | 5.5± 1.6 | 3.0 ± 13.5 | 23.4 ± 16.7 | 100.0 ± 4.2 | 0.0 ± 3.5 |
| Pediococcus sp. | 32.7 ± 6.0 | 24.3 ± 19.8 | 3.0 ± 9.3 | 2.7 ± 15.4 | −2.2 ± 5.5 | −3.0 ± 3.2 | 100.0 ± 6.4 | 0.0 ± 3.6 |
| Brettanomyces bruxellensis | 26.1 ± 17.4 | 27.4 ± 4.0 | 22.6 ± 6.2 | 2.5 ± 3.5 | 6.1 ± 15.4 | 11.0 ± 3.7 | 100.0 ± 13.8 | 0.0 ± 2.3 |
| Candida zeylanoides | 99.8 ± 6.7 | 73.1 ± 2.5 | 77.4 ± 10.2 | 73.9 ± 14.2 | 53.1 ± 17.7 | 21.8 ± 9.8 | 100.0 ± 27.7 | 0.0 ± 3.5 |
| Hanseniaspora uvarum | 85.7 ± 0.6 | 46.8 ± 14.2 | 5.9 ± 1.9 | 8.4 ± 3.4 | 8.4 ± 22.4 | 5.0 ± 3.0 | 100.0 ± 7.6 | 0.0 ± 5.9 |
| Metschnikowia pulcherrima | 56.0 ± 14.0 | 74.6 ± 8.9 | 67.3 ± 7.6 | 72.3 ± 6.8 | 39.6 ± 19.7 | 28.2 ± 22.1 | 100.0 ± 3.3 | 0.0 ± 1.5 |
| Pichia fermentans | 96.3 ± 10.6 | 91.2 ± 0.8 | 83.6 ± 0.1 | 81.1 ± 2.5 | 25.0 ± 26.3 | 7.7 ± 11.6 | 100.0 ± 2.8 | 0.0 ± 0.6 |
| Saccharomyces cerevisiae | 29.4 ± 6.1 | 3.8 ± 3.1 | 10.0 ± 18.5 | −0.9 ± 1.2 | 1.1 ± 4.1 | 7.3 ± 3.4 | 100.0 ± 3.1 | 0.0 ± 1.0 |
| Schizosaccharomyces pombe | 88.1 ± 2.4 | 79.0 ± 2.3 | 74.7 ± 2.0 | 67.5 ± 3.7 | 31.9 ± 26.0 | 24.0 ± 33.4 | 100.0 ± 2.9 | 0.0 ± 3.6 |
| Torulaspora delbruckii | 84.4 ± 5.2 | 86.4 ± 8.1 | 93.4 ± 4.6 | 5.5 ± 5.7 | 4.9 ± 1.4 | 15.3 ± 4.2 | 100.0 ± 2.2 | 0.0 ± 1.1 |
| Zygosaccharomyces bailii | 85.1 ± 16.1 | 84.3 ± 4.8 | 79.3 ± 8.5 | 66.2 ± 2.0 | 13.2 ± 6.7 | 40.1 ± 7.7 | 100.0 ± 43.4 | 0.0 ± 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stahl, L.F.; Edo, M.; Nonnenmacher, T.; Reif, D.; Rex, F.; Wegmann-Herr, P.; Kortekamp, A.; Fischer-Schuch, J.; Thines, E.; Scharfenberger-Schmeer, M. Ozonized Oleic Acid as a New Viticultural Treatment? Study of the Effect of LIQUENSO® Oxygenate on the Carpoplane Microbial Community and Wine Microorganisms Combining Metabarcoding and In Vitro Assays. Ecologies 2022, 3, 292-307. https://doi.org/10.3390/ecologies3030023
Stahl LF, Edo M, Nonnenmacher T, Reif D, Rex F, Wegmann-Herr P, Kortekamp A, Fischer-Schuch J, Thines E, Scharfenberger-Schmeer M. Ozonized Oleic Acid as a New Viticultural Treatment? Study of the Effect of LIQUENSO® Oxygenate on the Carpoplane Microbial Community and Wine Microorganisms Combining Metabarcoding and In Vitro Assays. Ecologies. 2022; 3(3):292-307. https://doi.org/10.3390/ecologies3030023
Chicago/Turabian StyleStahl, Lea Franziska, Manon Edo, Timon Nonnenmacher, Daniela Reif, Friederike Rex, Pascal Wegmann-Herr, Andreas Kortekamp, Jochen Fischer-Schuch, Eckhard Thines, and Maren Scharfenberger-Schmeer. 2022. "Ozonized Oleic Acid as a New Viticultural Treatment? Study of the Effect of LIQUENSO® Oxygenate on the Carpoplane Microbial Community and Wine Microorganisms Combining Metabarcoding and In Vitro Assays" Ecologies 3, no. 3: 292-307. https://doi.org/10.3390/ecologies3030023
APA StyleStahl, L. F., Edo, M., Nonnenmacher, T., Reif, D., Rex, F., Wegmann-Herr, P., Kortekamp, A., Fischer-Schuch, J., Thines, E., & Scharfenberger-Schmeer, M. (2022). Ozonized Oleic Acid as a New Viticultural Treatment? Study of the Effect of LIQUENSO® Oxygenate on the Carpoplane Microbial Community and Wine Microorganisms Combining Metabarcoding and In Vitro Assays. Ecologies, 3(3), 292-307. https://doi.org/10.3390/ecologies3030023






