Exploring the Bottleneck in Cryo-EM Dynamic Disorder Feature and Advanced Hybrid Prediction Model
Abstract
1. Introduction
2. Methods
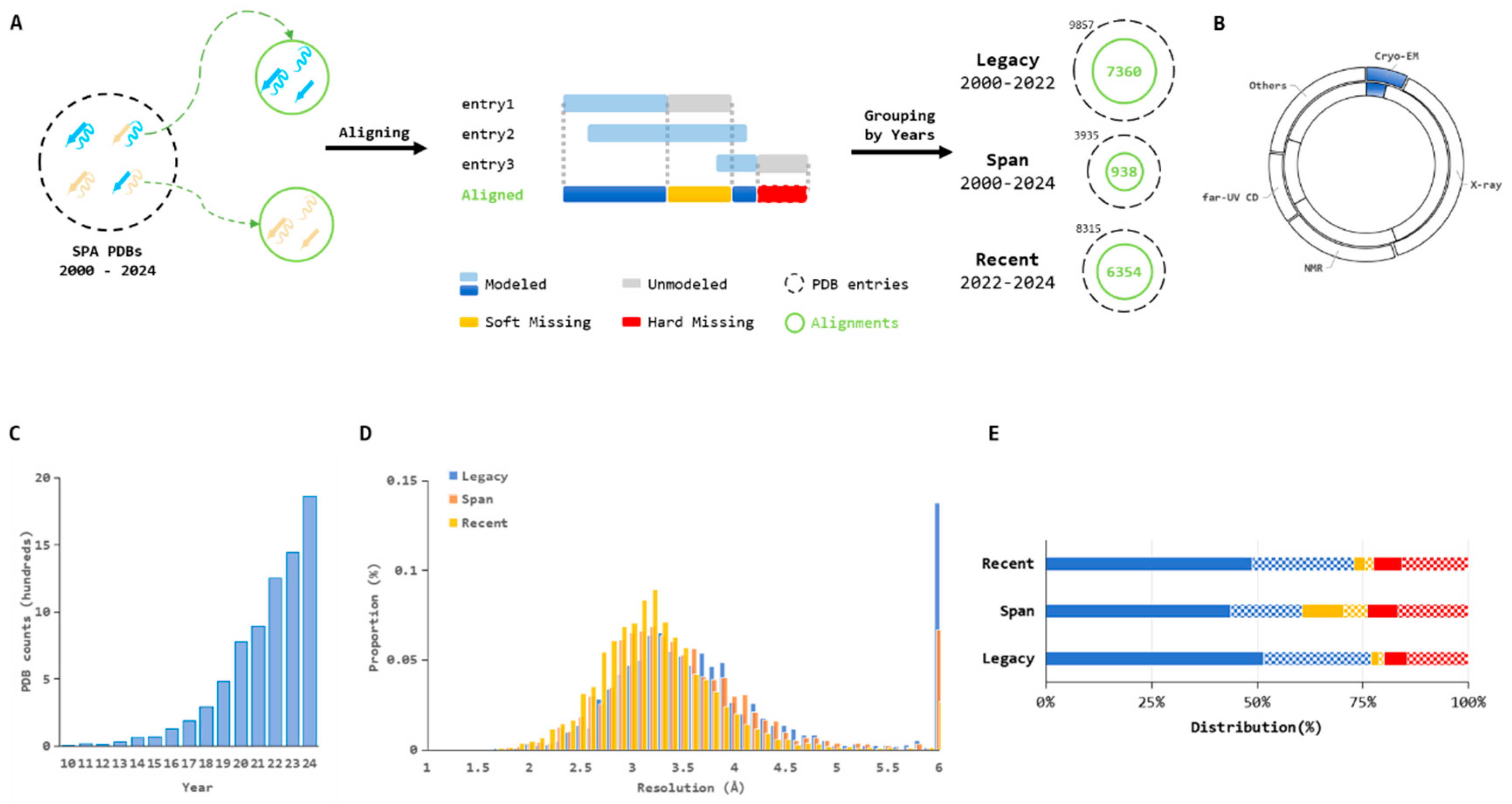
2.1. Data Preparation from Cryo-EM Single-Particle Analysis
- Legacy: This category includes all aligned entries deposited between 2000 and 2022, representing historical structural data (Training dataset).
- Span: This category comprises aligned entries that span the entire period from 2000 to 2024, capturing datasets that have been continuously updated over time (Validation dataset 01).
- Recent: This category consists of aligned entries deposited between 2022 and 2024, reflecting the latest advancements in SPA (Validation dataset 02).
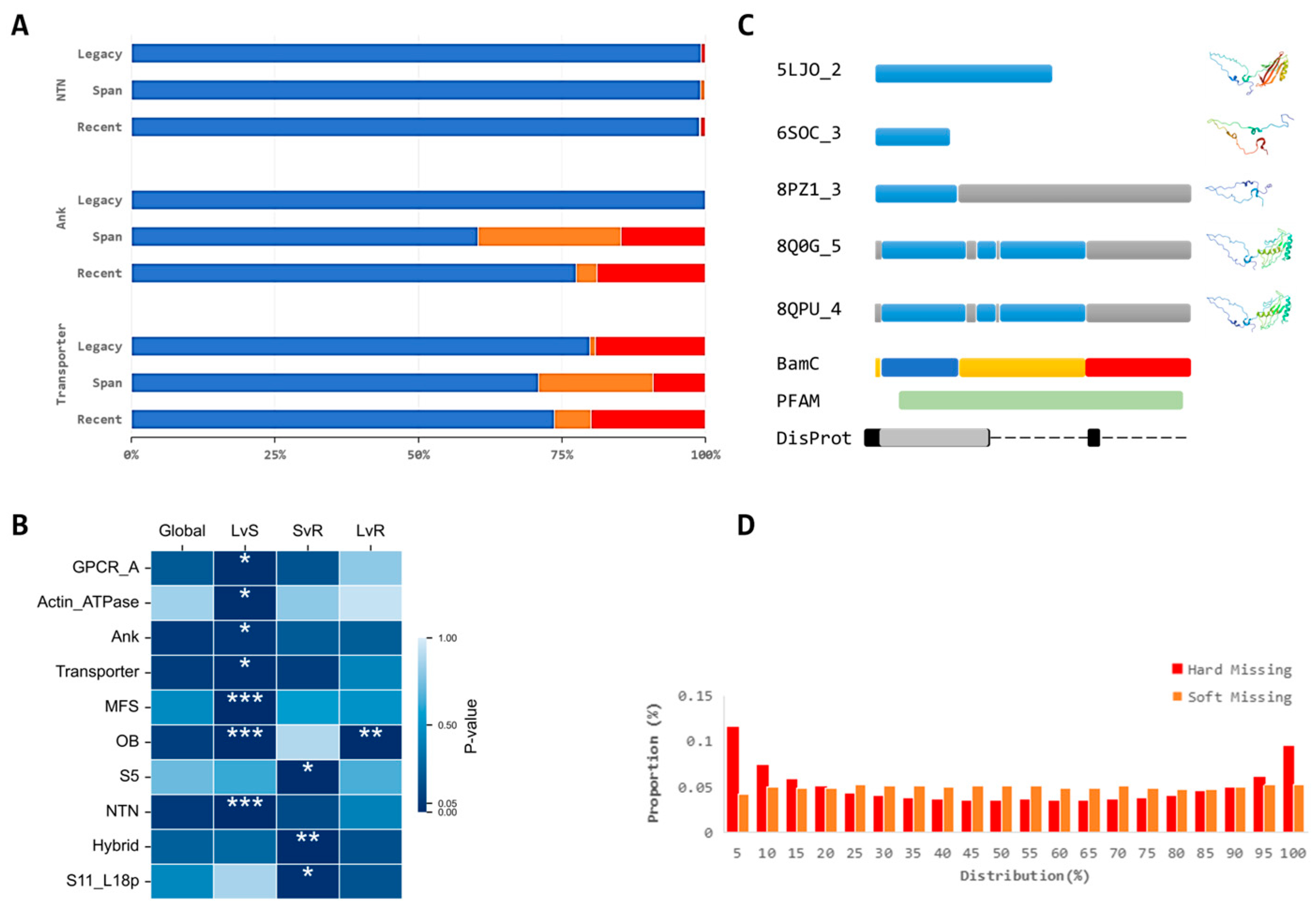
2.2. Structural Absence Classification, Functional Annotation, and Statical Analysis
- (1)
- Modeled: Residues with Cα coordinates consistently present in all corresponding PDB entities at the same residue location.
- (2)
- Soft Missing: Residues with Cα coordinates present in some, but not all, corresponding PDB entities.
- (3)
- Hard Missing: Residues entirely lacking Cα coordinates in all corresponding PDB entities.
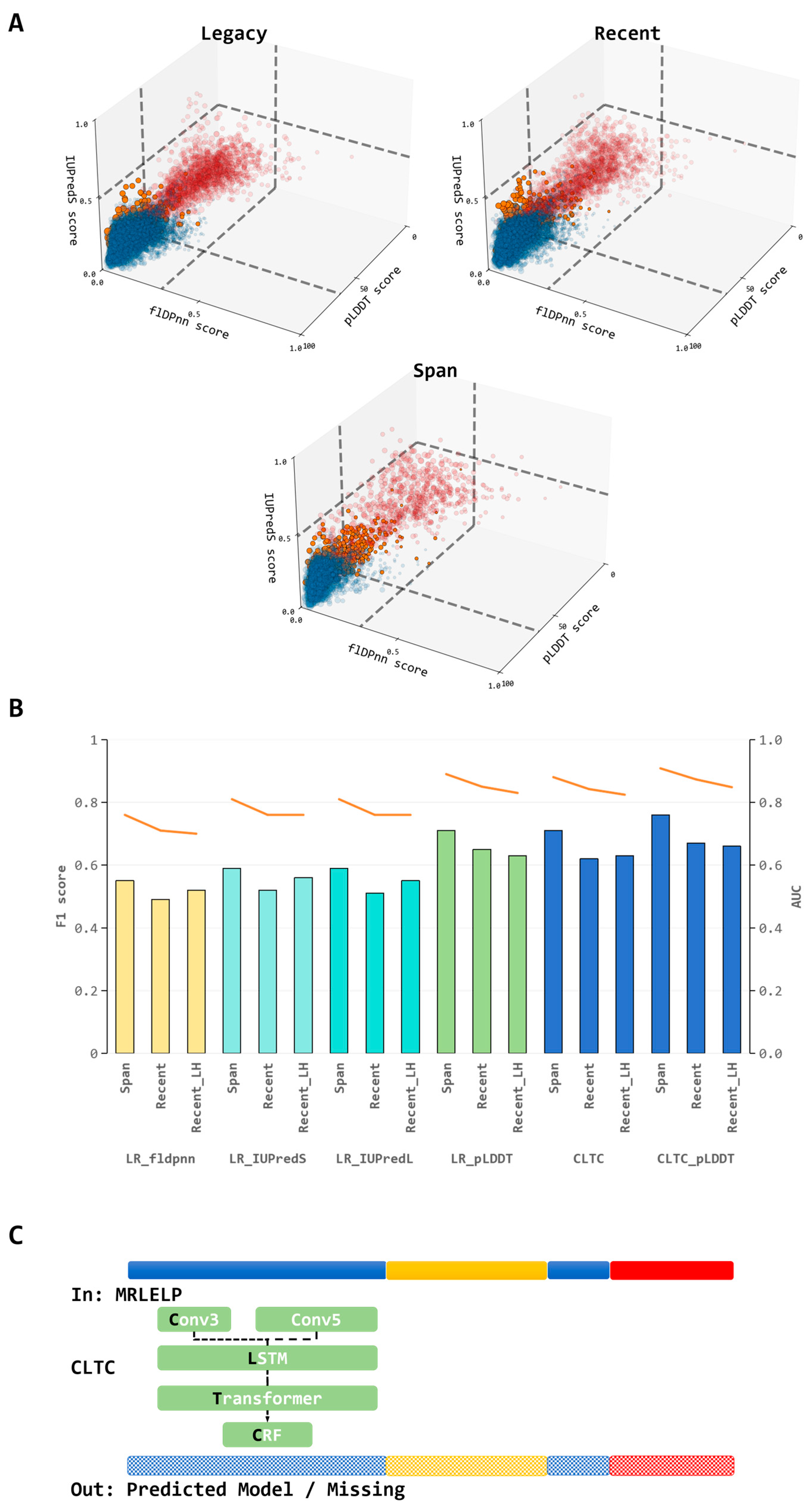
2.3. Disorder Feature Calculation and Calibration with Structure Absence
- (1)
- pLDDT: Values were retrieved from downloaded AlphaFold2 predicted structures from the AlphaFoldDB (AlphaFold version 2.0, https://alphafold.ebi.ac.uk/).
- (2)
- flDPnn: Values were generated using the Docker implementation of flDPnn [19] with default setting (flDPnn docker version December 2021, https://gitlab.com/sina.ghadermarzi/fldpnn_docker, accessed on 13 July 2025).
- (3)
- IUPred: Values were computed using the latest version of IUPred3 [17] in either “Short” (IUPredS) or “Long” (IUPredL) modes with default settings (version 3.0, https://iupred3.elte.hu/download_new, accessed on 13 July 2025).
2.4. Hybrid Model Architecture and Optimization
2.5. Verification of Model Predictions
- (1)
- True positives (TPs): A residue is correctly predicted as belonging to its respective category.
- (2)
- True negatives (TNs): A residue is correctly predicted as not belonging to a specific category.
- (3)
- False positives (FPs): A residue is incorrectly predicted as belonging to a specific category when it does not.
- (4)
- False negatives (FNs): A residue is wrongly predicted as not belonging to a specific category when it actually does.
2.6. Software
3. Results
3.1. Temporal Analyses of Dynamic Maps in Cryo-EM Single-Particle Analysis
3.2. Characteristics of Structural Absences in Single-Particle Analysis
3.3. Variable Performance of Disorder Prediction Tools in Characterizing Cryo-EM Structural Absences
3.4. Predictive Performance of the Hybrid Model and Advancement
4. Discussion
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuhlbrandt, W. The resolution revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef] [PubMed]
- Hagen, W.J.H.; Wan, W.; Briggs, J.A.G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 2017, 197, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Cressey, D.; Callaway, E. Cryo-electron microscopy wins chemistry Nobel. Nature 2017, 550, 167. [Google Scholar] [CrossRef] [PubMed]
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-particle cryo-EM at atomic resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Obradovic, Z.; Smith, D.K.; Zhu, G.; Vucetic, S.; Brown, C.J.; Lawson, J.D.; Dunker, A.K. Protein flexibility and intrinsic disorder. Protein Sci. 2004, 13, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, T.; Romero, P.R.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in the Protein Data Bank. J. Biomol. Struct. Dyn. 2007, 24, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef] [PubMed]
- Receveur-Brechot, V.; Bourhis, J.M.; Uversky, V.N.; Canard, B.; Longhi, S. Assessing protein disorder and induced folding. Proteins 2006, 62, 24–45. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Futschik, M.E.; Teichmann, S.A.; Babu, M.M. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science 2008, 322, 1365–1368. [Google Scholar] [CrossRef] [PubMed]
- Nwanochie, E.; Uversky, V.N. Structure Determination by Single-Particle Cryo-Electron Microscopy: Only the Sky (and Intrinsic Disorder) is the Limit. Int. J. Mol. Sci. 2019, 20, 4186. [Google Scholar] [CrossRef] [PubMed]
- Dodd, T.; Yan, C.; Ivanov, I. Simulation-Based Methods for Model Building and Refinement in Cryoelectron Microscopy. J. Chem. Inf. Model. 2020, 60, 2470–2483. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically Disordered Proteins and Their “Mysterious” (Meta)Physics. Front. Phys. 2019, 7, 10. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Erdos, G.; Dosztanyi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Erdos, G.; Pajkos, M.; Dosztanyi, Z. IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef] [PubMed]
- Hatos, A.; Hajdu-Soltesz, B.; Monzon, A.M.; Palopoli, N.; Alvarez, L.; Aykac-Fas, B.; Bassot, C.; Benitez, G.I.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020, 48, D269–D276. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Zidek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Saldano, T.; Escobedo, N.; Marchetti, J.; Zea, D.J.; Mac Donagh, J.; Velez Rueda, A.J.; Gonik, E.; Garcia Melani, A.; Novomisky Nechcoff, J.; Salas, M.N.; et al. Impact of protein conformational diversity on AlphaFold predictions. Bioinformatics 2022, 38, 2742–2748. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.J.; Choy, W.Y.; Karttunen, M. AlphaFold2: A Role for Disordered Protein/Region Prediction? Int. J. Mol. Sci. 2022, 23, 4591. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Kurgan, L. Surveying over 100 predictors of intrinsic disorder in proteins. Expert. Rev. Proteom. 2021, 18, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Necci, M.; Piovesan, D.; Predictors, C.; DisProt, C.; Tosatto, S.C.E. Critical assessment of protein intrinsic disorder prediction. Nat. Methods 2021, 18, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Sun, F. Developing Graphene Grids for Cryoelectron Microscopy. Front. Mol. Biosci. 2022, 9, 937253. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wang, H.W. Better Cryo-EM Specimen Preparation: How to Deal with the Air-Water Interface? J. Mol. Biol. 2023, 435, 167926. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Lin, P.; Chen, J.; Cao, H.; Huang, S.Y. Model building of protein complexes from intermediate-resolution cryo-EM maps with deep learning-guided automatic assembly. Nat. Commun. 2022, 13, 4066. [Google Scholar] [CrossRef] [PubMed]
- Jamali, K.; Kall, L.; Zhang, R.; Brown, A.; Kimanius, D.; Scheres, S.H.W. Automated model building and protein identification in cryo-EM maps. Nature 2024, 628, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Arseni, D.; Zhang, W.; Huang, M.; Lovestam, S.; Schweighauser, M.; Kotecha, A.; Murzin, A.G.; Peak-Chew, S.Y.; Macdonald, J.; et al. Cryo-EM structures of amyloid-beta 42 filaments from human brains. Science 2022, 375, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Aspromonte, M.C.; Nugnes, M.V.; Quaglia, F.; Bouharoua, A.; DisProt, C.; Tosatto, S.C.E.; Piovesan, D. DisProt in 2024: Improving function annotation of intrinsically disordered proteins. Nucleic Acids Res. 2024, 52, D434–D441. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S. Navigating the unstructured by evaluating alphafold’s efficacy in predicting missing residues and structural disorder in proteins. PLoS ONE 2025, 20, e0313812. [Google Scholar] [CrossRef] [PubMed]
- Seoane, B.; Carbone, A. Soft disorder modulates the assembly path of protein complexes. PLoS Comput. Biol. 2022, 18, e1010713. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Soding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Barrio-Hernandez, I.; Yeo, J.; Janes, J.; Mirdita, M.; Gilchrist, C.L.M.; Wein, T.; Varadi, M.; Velankar, S.; Beltrao, P.; Steinegger, M. Clustering predicted structures at the scale of the known protein universe. Nature 2023, 622, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef] [PubMed]
- DeForte, S.; Uversky, V.N. Resolving the ambiguity: Making sense of intrinsic disorder when PDB structures disagree. Protein Sci. 2016, 25, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Hagan, C.L.; Kim, S.; Kahne, D. Reconstitution of outer membrane protein assembly from purified components. Science 2010, 328, 890–892. [Google Scholar] [CrossRef] [PubMed]
- Iadanza, M.G.; Higgins, A.J.; Schiffrin, B.; Calabrese, A.N.; Brockwell, D.J.; Ashcroft, A.E.; Radford, S.E.; Ranson, N.A. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat. Commun. 2016, 7, 12865. [Google Scholar] [CrossRef] [PubMed]
- Fenn, K.L.; Horne, J.E.; Crossley, J.A.; Bohringer, N.; Horne, R.J.; Schaberle, T.F.; Calabrese, A.N.; Radford, S.E.; Ranson, N.A. Outer membrane protein assembly mediated by BAM-SurA complexes. Nat. Commun. 2024, 15, 7612. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.X.; Kalmar, L.; Tompa, P.; Han, K.H.; Selenko, P.; Dunker, A.K.; Daughdrill, G.W.; Uversky, V.N. The alphabet of intrinsic disorder: I. Act like a Pro: On the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disord. Proteins 2013, 1, e24360. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Kurgan, L. Compositional Bias of Intrinsically Disordered Proteins and Regions and Their Predictions. Biomolecules 2022, 12, 888. [Google Scholar] [CrossRef] [PubMed]
- Kurgan, L.; Hu, G.; Wang, K.; Ghadermarzi, S.; Zhao, B.; Malhis, N.; Erdos, G.; Gsponer, J.; Uversky, V.N.; Dosztanyi, Z. Tutorial: A guide for the selection of fast and accurate computational tools for the prediction of intrinsic disorder in proteins. Nat. Protoc. 2023, 18, 3157–3172. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Porter, L.L. AlphaFold2 fails to predict protein fold switching. Protein Sci. 2022, 31, e4353. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Liebschner, D.; Croll, T.I.; Williams, C.J.; McCoy, A.J.; Poon, B.K.; Afonine, P.V.; Oeffner, R.D.; Richardson, J.S.; Read, R.J.; et al. AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination. Nat. Methods 2024, 21, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; McShan, A.C. The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins. Nat. Chem. Biol. 2024, 20, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.; Paliwal, K.K.; Litfin, T.; Zhou, Y. SPOT-Disorder2: Improved Protein Intrinsic Disorder Prediction by Ensembled Deep Learning. Genom. Proteom. Bioinform. 2019, 17, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.; Paliwal, K.; Zhou, Y. Accurate Single-Sequence Prediction of Protein Intrinsic Disorder by an Ensemble of Deep Recurrent and Convolutional Architectures. J. Chem. Inf. Model. 2018, 58, 2369–2376. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Mahmoud, Q.H. Design and Development of RNN Anomaly Detection Model for IoT Networks. IEEE Access 2022, 10, 62722–62750. [Google Scholar] [CrossRef]
- Dou, B.; Zhu, Z.; Merkurjev, E.; Ke, L.; Chen, L.; Jiang, J.; Zhu, Y.; Liu, J.; Zhang, B.; Wei, G.W. Machine Learning Methods for Small Data Challenges in Molecular Science. Chem. Rev. 2023, 123, 8736–8780. [Google Scholar] [CrossRef] [PubMed]
- Colon-Ruiz, C.; Segura-Bedmar, I. Comparing deep learning architectures for sentiment analysis on drug reviews. J. Biomed. Inform. 2020, 110, 103539. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Outeiral, C.; Nissley, D.A.; Deane, C.M. Current structure predictors are not learning the physics of protein folding. Bioinformatics 2022, 38, 1881–1887. [Google Scholar] [CrossRef] [PubMed]
- Alderson, T.R.; Pritisanac, I.; Kolaric, D.; Moses, A.M.; Forman-Kay, J.D. Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2. Proc. Natl. Acad. Sci. USA 2023, 120, e2304302120. [Google Scholar] [CrossRef] [PubMed]
| Dataset | Models | Precision | Recall | F1 | AUC |
|---|---|---|---|---|---|
| Span | L | 0.78 | 0.52 | 0.63 | 0.86 |
| T | 0.39 | 0.58 | 0.47 | 0.70 | |
| LT | 0.71 | 0.64 | 0.67 | 0.85 | |
| CLT | 0.72 | 0.66 | 0.69 | 0.87 | |
| CLC | 0.69 | 0.67 | 0.68 | 0.86 | |
| CTC | 0.46 | 0.65 | 0.54 | 0.77 | |
| CLTC | 0.74 | 0.67 | 0.71 | 0.88 | |
| Recent | L | 0.77 | 0.42 | 0.54 | 0.82 |
| T | 0.38 | 0.59 | 0.46 | 0.71 | |
| LT | 0.63 | 0.57 | 0.60 | 0.83 | |
| CLT | 0.61 | 0.61 | 0.61 | 0.84 | |
| CLC | 0.63 | 0.58 | 0.60 | 0.82 | |
| CTC | 0.45 | 0.59 | 0.51 | 0.76 | |
| CLTC | 0.62 | 0.62 | 0.62 | 0.84 | |
| Recent_LH | L | 0.77 | 0.44 | 0.56 | 0.80 |
| T | 0.42 | 0.61 | 0.50 | 0.71 | |
| LT | 0.65 | 0.57 | 0.61 | 0.81 | |
| CLT | 0.63 | 0.60 | 0.62 | 0.82 | |
| CLC | 0.64 | 0.60 | 0.62 | 0.82 | |
| CTC | 0.47 | 0.59 | 0.51 | 0.76 | |
| CLTC | 0.63 | 0.63 | 0.63 | 0.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S. Exploring the Bottleneck in Cryo-EM Dynamic Disorder Feature and Advanced Hybrid Prediction Model. Biophysica 2025, 5, 39. https://doi.org/10.3390/biophysica5030039
Zheng S. Exploring the Bottleneck in Cryo-EM Dynamic Disorder Feature and Advanced Hybrid Prediction Model. Biophysica. 2025; 5(3):39. https://doi.org/10.3390/biophysica5030039
Chicago/Turabian StyleZheng, Sen. 2025. "Exploring the Bottleneck in Cryo-EM Dynamic Disorder Feature and Advanced Hybrid Prediction Model" Biophysica 5, no. 3: 39. https://doi.org/10.3390/biophysica5030039
APA StyleZheng, S. (2025). Exploring the Bottleneck in Cryo-EM Dynamic Disorder Feature and Advanced Hybrid Prediction Model. Biophysica, 5(3), 39. https://doi.org/10.3390/biophysica5030039





