Emergence of Diverse Epidermal Patterns via the Integration of the Turing Pattern Model with the Majority Voting Model
Abstract
1. Introduction
2. Models and Methods
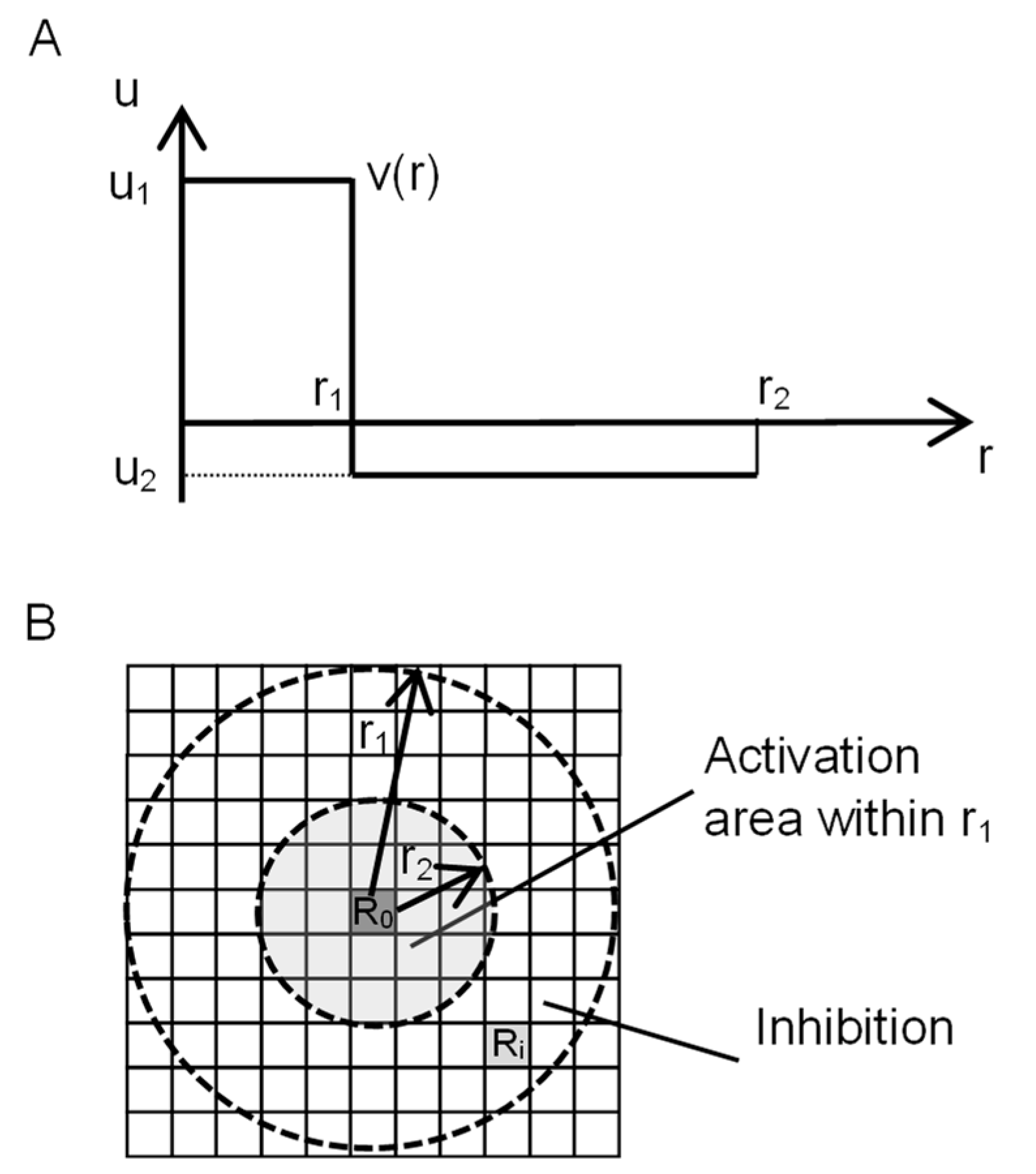
2.1. Overview of the Turing Pattern Model
2.2. Representation of the Turing Model Using CA
2.3. Majority Voting Model Using CA
2.4. Proposed Integration Model
2.5. Model with Invariant Regions at the Boundary of the Patterns
2.6. Calculation Conditions
- -
- Calculation field: 100 × 100 cells in a hexagonal grid;
- -
- Periodic boundary condition;
- -
- Initial conditions: states 0 and 1 were placed randomly in each cell of the computational field with a probability of 0.5;
- -
- At each time step, the cells in the lattice space were synchronously changed, and the computation was repeated until the pattern formation stabilized;
- -
- The range of S1 was set to three cells from the focal cell, and the range of S2 was set to six cells from the focal cell. The parameter s determines the scale of the pattern to be created and, if it is larger, the patterns will only become more similar and larger. For this reason, s was fixed.
3. Results
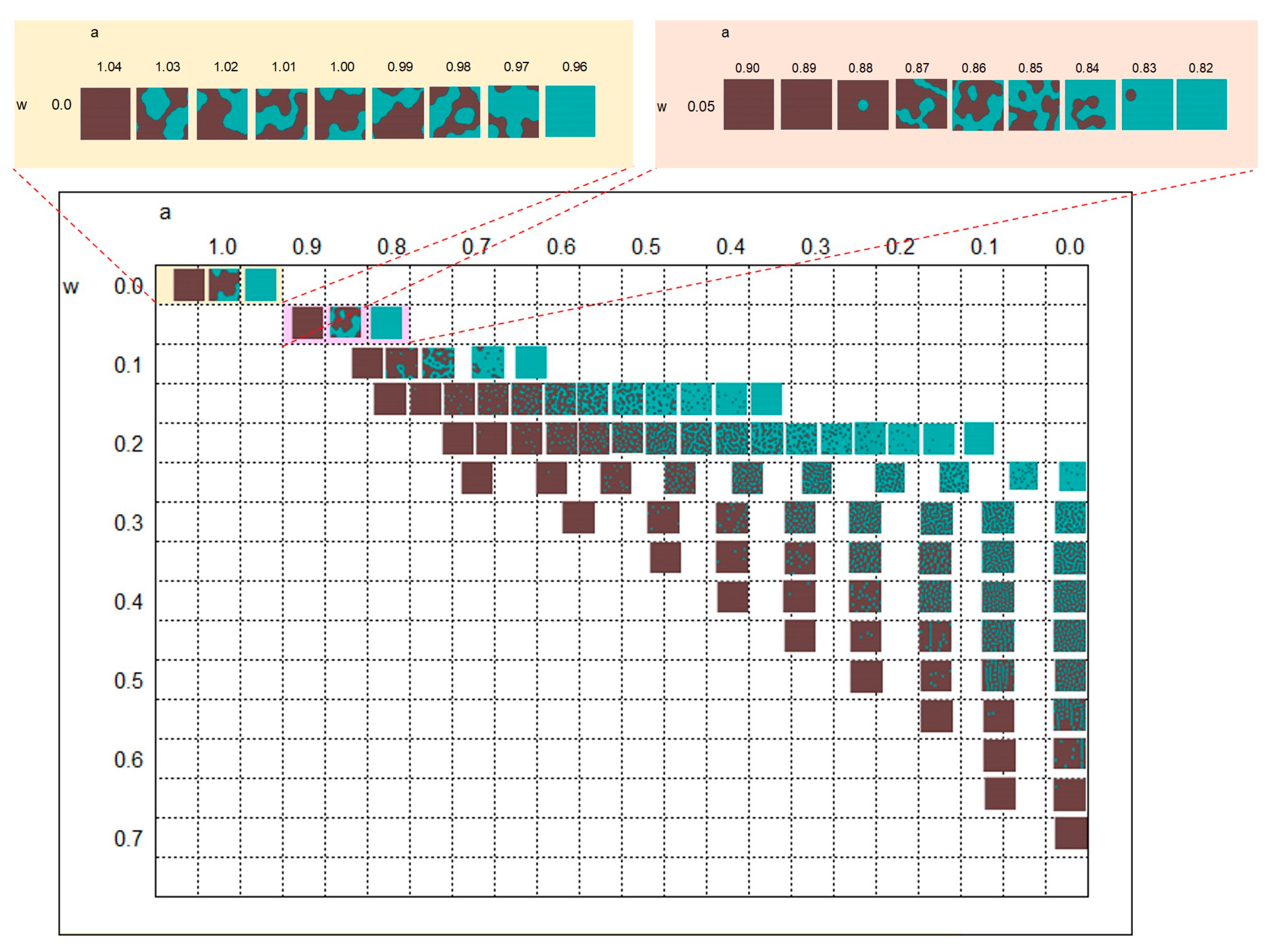
3.1. Parameter Map
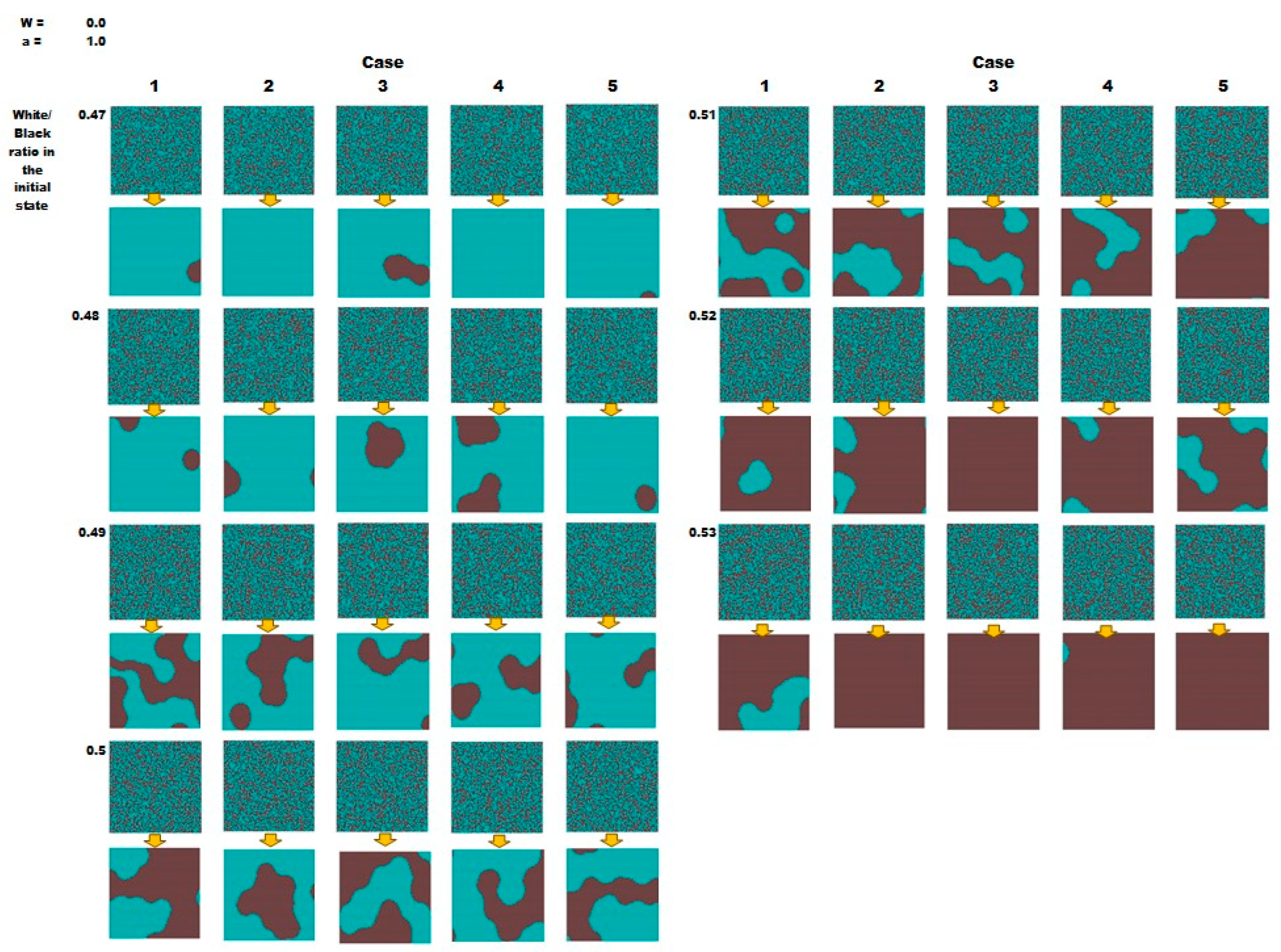
3.2. Initial Value Dependency
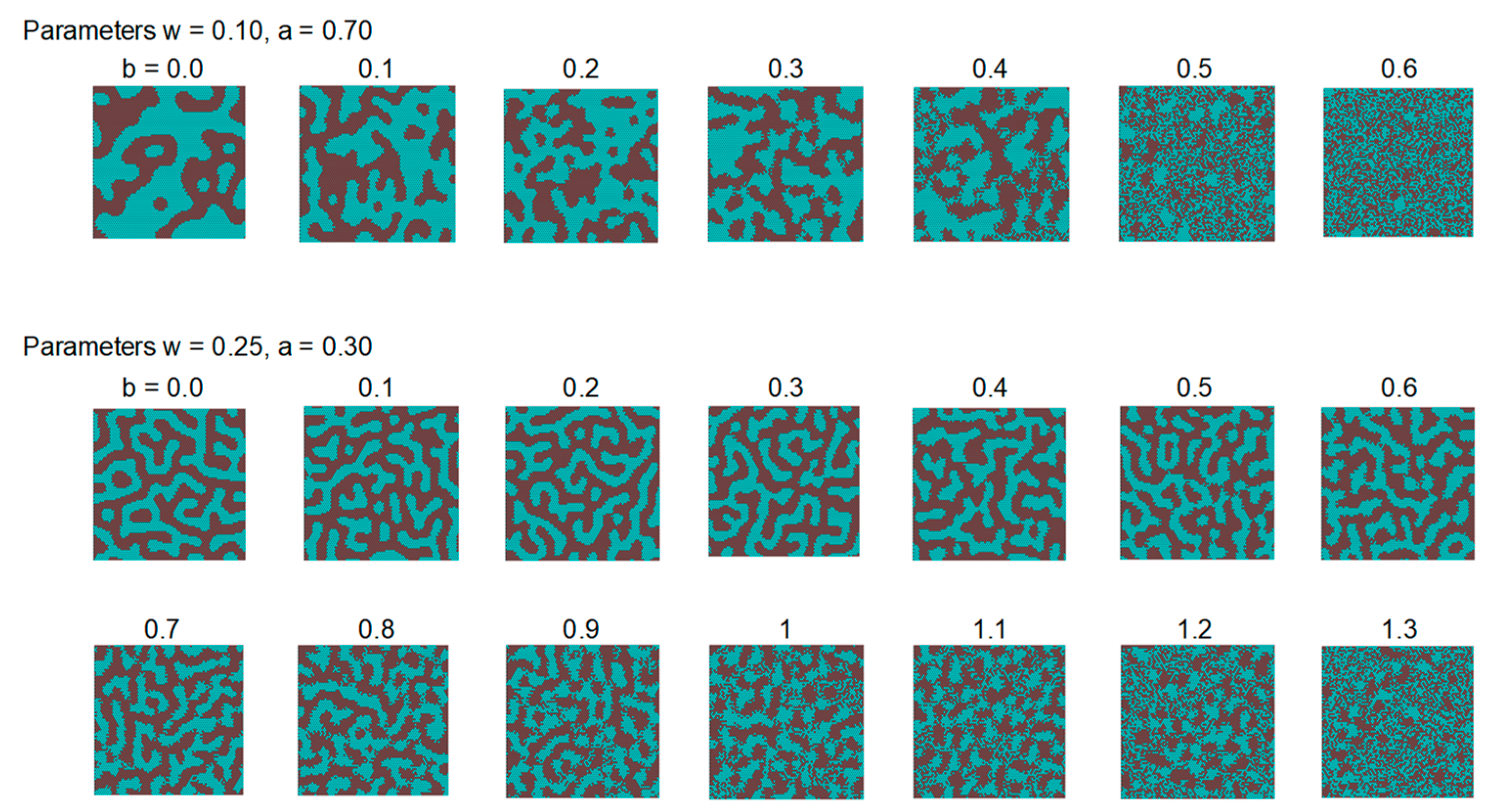
3.3. Results of the Model with Invariant Regions at the Boundary of the Patterns
4. Discussion
5. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turing, A.M. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 1952, 237, 37–72. [Google Scholar]
- Meinhardt, H. Models of Biological Pattern Formation; Academic Press: New York, NY, USA, 1982; Volume 6. [Google Scholar]
- Wolpert, L. Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 1969, 25, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Wolpert, L. Positional information revisited. Development 1989, 107 (Suppl. S3), 3–12. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Asal, R. A reaction-diffusion wave on the skin of the marine angelfish Pomacanthus. Nature 1995, 376, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, S.; Okamoto, M.; Kondo, S. Blending of animal colour patterns by hybridization. Nat. Commun. 2010, 1, 66. [Google Scholar] [CrossRef] [PubMed]
- Milinkovitch, M.C.; Jahanbakhsh, E.; Zakany, S. The unreasonable effectiveness of reaction diffusion in vertebrate skin color patterning. Annu. Rev. Cell Dev. Biol. 2023, 39, 145–174. [Google Scholar] [CrossRef] [PubMed]
- Entchev, E.V.; Schwabedissen, A.; González-Gaitán, M. Gradient formation of the TGF-beta homolog Dpp. Cell 2000, 103, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Sick, S.; Reinker, S.; Timmer, J.; Schlake, T. WNT and DKK determine hair follicle spacing through a reaction-diffusion mechanism. Science 2006, 314, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Economou, A.D.; Ohazama, A.; Porntaveetus, T.; Sharpe, P.T.; Kondo, S.; Basson, M.A.; Gritli-Linde, A.; Cobourne, M.T.; Green, J.B.A. Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat. Genet. 2012, 44, 348–351. [Google Scholar] [CrossRef]
- Dilão, R.; Muraro, D. mRNA diffusion explains protein gradients in Drosophila early development. J. Theor. Biol. 2010, 264, 847–853. [Google Scholar] [CrossRef]
- Dilão, R. Bicoid mRNA diffusion as a mechanism of morphogenesis in Drosophila early development. Comptes Rendus Biol. 2014, 337, 679–682. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Yoshimoto, E.; Kondo, S. Pattern regulation in the stripe of zebrafish suggests an underlying dynamic and autonomous mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 4790–4793. [Google Scholar] [CrossRef]
- Nakamasu, A.; Takahashi, G.; Kanbe, A.; Kondo, S. Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc. Natl. Acad. Sci. USA 2009, 106, 8429–8434. [Google Scholar] [CrossRef]
- Frohnhöfer, H.G.; Krauss, J.; Maischein, H.M.; Nüsslein-Volhard, C. Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development 2013, 140, 2997–3007. [Google Scholar] [CrossRef]
- Woolley, T.E. Pattern production through a chiral chasing mechanism. Phys. Rev. E 2017, 96, 032401. [Google Scholar] [CrossRef]
- Yamanaka, H.; Kondo, S. In vitro analysis suggests that difference in cell movement during direct interaction can generate various pigment patterns in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 1867–1872. [Google Scholar] [CrossRef]
- Inaba, M.; Yamanaka, H.; Kondo, S. Pigment pattern formation by contact-dependent depolarization. Science 2012, 335, 677. [Google Scholar] [CrossRef]
- Hamada, H.; Watanabe, M.; Lau, H.E.; Nishida, T.; Hasegawa, T.; Parichy, D.M.; Kondo, S. Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development 2014, 141, 318–324. [Google Scholar] [CrossRef]
- Kondo, S.; Watanabe, M.; Miyazawa, S. Studies of Turing pattern formation in zebrafish skin. Philos. Trans. A Math. Phys. Eng. Sci. 2021, 379, 20200274. [Google Scholar] [CrossRef]
- Watanabe, M.; Kondo, S. Is pigment patterning in fish skin determined by the Turing mechanism? Trends Genet. 2015, 31, 88–96. [Google Scholar] [CrossRef]
- Bullara, D.; De Decker, Y. Pigment cell movement is not required for generation of Turing patterns in zebrafish skin. Nat. Commun. 2015, 6, 6971. [Google Scholar] [CrossRef]
- Caicedo-Carvajal, C.E.; Shinbrot, T. In silico zebrafish pattern formation. Dev. Biol. 2008, 315, 397–403. [Google Scholar] [CrossRef]
- Volkening, A.; Sandstede, B. Modelling stripe formation in zebrafish: An agent-based approach. J. R. Soc. Interface 2015, 12, 20150812. [Google Scholar] [CrossRef]
- Volkening, A.; Sandstede, B. Iridophores as a source of robustness in zebrafish stripes and variability in Danio patterns. Nat. Commun. 2018, 9, 3231. [Google Scholar] [CrossRef]
- Vasilopoulos, G.; Painter, K.J. Pattern formation in discrete cell tissues under long range filopodia-based direct cell to cell contact. Math. Biosci. 2016, 273, 1–15. [Google Scholar] [CrossRef]
- Marcon, L.; Diego, X.; Sharpe, J.; Müller, P. High-throughput mathematical analysis identifies Turing networks for patterning with equally diffusing signals. eLife 2016, 5, e14022. [Google Scholar] [CrossRef]
- Moreira, J.; Deutsch, A. Pigment pattern formation in zebrafish during late larval stages: A model based on local interactions. Dev. Dyn. 2005, 232, 33–42. [Google Scholar] [CrossRef]
- Konow, C.; Li, Z.; Shepherd, S.; Bullara, D.; Epstein, I.R. Influence of survival, promotion, and growth on pattern formation in zebrafish skin. Sci. Rep. 2021, 11, 9864. [Google Scholar] [CrossRef]
- Owen, J.P.; Kelsh, R.N.; Yates, C.A. A quantitative modelling approach to zebrafish pigment pattern formation. eLife 2020, 9, e52998. [Google Scholar] [CrossRef]
- Ishida, T.; Tadokoro, H.; Takahashi, H.; Yoshikawa, H.; Harumi, S. Constructing models to reproduce the skin color patterns of Takifugu species, including hybrids, fisheries. Engineering 2019, 15, 15–26. [Google Scholar]
- Zakany, S.; Smirnov, S.; Milinkovitch, M.C. Lizard skin patterns and the Ising model. Phys. Rev. Lett. 2022, 128, 048102. [Google Scholar] [CrossRef]
- Graner, F.; Glazier, J.A. Simulation of biological cell sorting using a two-dimensional extended Potts model. Phys. Rev. Lett. 1992, 69, 2013–2016. [Google Scholar] [CrossRef]
- Dilão, R.; Sainhas, J. Modelling butterfly wing eyespot patterns. Proc. Biol. Sci. 2004, 271, 1565–1569. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S. An updated kernel-based Turing model for studying the mechanisms of biological pattern formation. J. Theor. Biol. 2017, 414, 120–127. [Google Scholar] [CrossRef]
- Miyazawa, S. Pattern blending enriches the diversity of animal colorations. Sci. Adv. 2020, 6, eabb9107. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, S.; Xu, J.; Feng, J.; Mahboob, S.; Al-Ghanim, K.A.; Sun, X.; Xu, P. Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS ONE 2014, 9, e108200. [Google Scholar] [CrossRef]
- Qiu, J. Epigenetics: Unfinished symphony. Nature 2006, 441, 143–145. [Google Scholar] [CrossRef]
- Vichniac, G.Y. Simulating physics with cellular automata. Phys. D Nonlinear Phenom. 1984, 10, 96–116. [Google Scholar] [CrossRef]
- Gierer, A.; Meinhardt, H.A. A Theory of biological pattern formation. Kybernetik 1972, 12, 30–39. [Google Scholar] [CrossRef]
- Gray, P.; Scott, S. Autocatalytic reactions in the isothermal continuous stirred tank reactor. Chem. Eng. Sci. 1983, 38, 29–43. [Google Scholar] [CrossRef]
- Schepers, H.E.; Markus, M. Two types of performance of an isotropic cellular automaton: Stationary (Turing) patterns and spiral waves. Physica A 1992, 188, 337–343. [Google Scholar] [CrossRef]
- Young, D.A.A. A Local activator-inhibitor model of vertebrate skin patterns. Math. Biosci. 1984, 72, 51–58. [Google Scholar] [CrossRef]
- Adamatzky, A.; Martínez, G.J.; Mora, J.C.S.T. Phenomenology of reaction-diffusion binary-state cellular automata. Int. J. Bifürcation Chaos 2006, 16, 2985–3005. [Google Scholar] [CrossRef]
- Dormann, S.; Deutsch, A.; Lawniczak, A.T. Fourier analysis of turing-like pattern formation in cellular automaton models. Future Gen. Comp. Syst. 2001, 17, 901–909. [Google Scholar] [CrossRef]
- Tsai, L.L.; Hutchison, G.R.; Peacock-López, E. Turing patterns in a self-replicating mechanism with a self-complementary template. J. Chem. Phys. 2000, 113, 2003–2006. [Google Scholar] [CrossRef]
- Manukyan, L.; Montandon, S.A.; Fofonjka, A.; Smirnov, S.; Milinkovitch, M.C. A living mesoscopic cellular automaton made of skin scales. Nature 2017, 544, 173–179. [Google Scholar] [CrossRef]
- Ishida, T. Possibility of controlling self-organized patterns with totalistic cellular automata consisting of both rules like game of life and rules producing Turing patterns. Micromachines 2018, 9, 339. [Google Scholar] [CrossRef]
- Guisoni, N.; Diambra, L. Transient Turing patterns in a morphogenetic model. Front. Phys. 2022, 10, 927152. [Google Scholar] [CrossRef]
- Maini, P.K.; Woolley, T.E.; Baker, R.E.; Gaffney, E.A.; Lee, S.S. Turing’s model for biological pattern formation and the robustness problem. Interface Focus. 2012, 2, 487–496. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishida, T. Emergence of Diverse Epidermal Patterns via the Integration of the Turing Pattern Model with the Majority Voting Model. Biophysica 2024, 4, 283-297. https://doi.org/10.3390/biophysica4020020
Ishida T. Emergence of Diverse Epidermal Patterns via the Integration of the Turing Pattern Model with the Majority Voting Model. Biophysica. 2024; 4(2):283-297. https://doi.org/10.3390/biophysica4020020
Chicago/Turabian StyleIshida, Takeshi. 2024. "Emergence of Diverse Epidermal Patterns via the Integration of the Turing Pattern Model with the Majority Voting Model" Biophysica 4, no. 2: 283-297. https://doi.org/10.3390/biophysica4020020
APA StyleIshida, T. (2024). Emergence of Diverse Epidermal Patterns via the Integration of the Turing Pattern Model with the Majority Voting Model. Biophysica, 4(2), 283-297. https://doi.org/10.3390/biophysica4020020







