Abstract
Bioluminescence resonance energy transfer (BRET) seems to be a promising biophysical technique to study protein–protein interactions within living cells due to a very specific reaction of bioluminescence that essentially decreases the background of other cellular components and light-induced destruction of biomacromolecules. An important direction of the development of this technique is the study of known strong protein–protein complexes in vivo and the estimation of an average distance between chromophores of the donor and acceptor. Here, we demonstrate an in vivo interaction between barnase fused with luciferase (from Renilla reniformis, RLuc) and barstar fused with EGFP (enhanced green fluorescent protein of Aequorea victoria) monitored by BRET. The distance between the luciferase and EGFP chromophores within the complex has been evaluated as equal to (56 ± 2) Å.
1. Introduction
Bioluminescence resonance energy transfer (BRET) is widely distributed in nature and provides color-shifted light emission of a number of marine and other organisms in the absence of an external light source [1,2]. This phenomenon takes place due to the proximity of two photo-proteins: luciferase (as the donor of bioluminescence) and some fluorescent protein (for example, GFP (green fluorescent protein) as the acceptor of bioluminescence) [3,4]. BRET efficiency depends on the spectral properties of the donor and acceptor, the mutual orientation of their chromophores, and the distance between them [5,6]. At present, BRET is widely used to monitor the proximity of biomolecules both in vivo and in vitro (see, e.g., [6,7,8]). In contrast to fluorescence resonance energy transfer (FRET), the bioluminescence emission is produced as a result of oxidation of the luciferase cofactor [9,10], which allows using BRET in the presence of intensive scattering, autofluorescence, and photobleaching. The heterological expression of luciferase and GFP-like genes fused with genes of target proteins provides an effective tool for in vivo monitoring of protein–protein interactions [7,8,11,12]. In this case, the length and composition of linkers connecting fused partners may have some effect on BRET efficiency. Thus, BRET efficiency in the monitoring of in vivo interacting proteins fused with photoproteins depends on several factors that are difficult to predict. However, BRET has an important advantage over FRET, i.e., the possibility to shift the equilibrium toward complex stabilization through increasing excess of the GFP-fused partner, because its fluorescence background is absent in the case of BRET. Therefore, BRET markers are required for the cases when the complex is predominately present or absent. In the present work, the in vivo interaction of barnase and barstar known to be very tight [13,14,15] has been probed by BRET. The well-known donor–acceptor pair containing photoproteins RLuc (luciferase from Renilla reniformis) [16,17,18,19,20,21] and GFP [4,21,22] fused with barnase and barstar correspondingly was used to monitor the BRET of the complex. The genes encoding the fusion proteins barnase–luciferase (RLuc) and barstar–EGFP (an enhanced variant of Aequoria victoria green fluorescent protein) were expressed in Escherichia coli cells both separately and together (co-expression). The expression of the fused barstar–EGFP gene was ten-fold higher than that of the fused barnase–luciferase gene, thus providing full saturation of the complex barnase–barstar. It was found that the expression of solely the barnase–luciferase gene results in poor cell growth and a low biosynthesis level of the fusion protein probably due to the high toxicity of barnase [23]. On the other hand, co-expression of both plasmids encoding fusion proteins barnase–luciferase and barstar–EGFP leads to the elimination of barnase toxicity and normal growth of the cells. Thus, the functional modules of the fusion proteins (barnase and barstar) have native conformations after biosynthesis in the cell. Moreover, the bioluminescence spectrum of the cells after co-expression of both fusion proteins contains the specific emission band of EGFP (at 510 nm) that confirms the presence of tight barnase–barstar complex in vivo providing the proximity of photoprotein chromophores by an evaluated distance of (56 ± 2) Å.
2. Materials and Methods
2.1. Materials
The following chemicals were used: NaCl, NaOH, HCl (Reachem, Moscow, Russia); Tris, acrylamide, ammonium sulfate, SDS, β-mercaptoethanol, marker proteins (Sigma-Aldrich, St. Louis, MO, USA); HEPES, KCl, DTT, MgCl2, CaCl2, Na2HPO4, NaH2PO4 (ICN Biomedicals, Costa Mesa, CA, USA); coelenterazine (Intrinsic Bioprobes, Tempe, AZ, USA). Restriction endonucleases, T4 ligase, dNTPs, and isopropyl β-D-1-thiogalactopyranoside (IPTG) were from Thermo Fisher Scientific (Vilnius, Lithuania). Pfu Turbo high fidelity DNA polymerase was purchased from AlfaFerment (Moscow, Russia).
2.2. Strains and Culture Conditions
Escherichia coli strains DH5α and BL21(DE3) were used to amplify plasmid DNA and to express genes, respectively. E. coli strains were routinely grown in lysogeny broth (LB) at 37 °C. For solid medium, LB was supplemented with Bacto agar (1.5%, w/v). When appropriate, kanamycin (20 µg/mL) and ampicillin (100 µg/mL) were added to the medium.
2.3. Genetic Manipulations
Standard procedures were used for restriction, ligation, gel electrophoresis, and transformation of E. coli cells [24]. Plasmid DNAs were purified using a QIAprep Spin Miniprep Kit (QIAGEN, Hilden, Germany), DNA fragments were purified from agarose gels with a QIAquick Gel extraction Kit (QIAGEN), and PCR fragments were purified with a QIAquick PCR purification Kit (QIAGEN). Co-expression of genes was facilitated by the use of the pair of plasmids pAC28 [25] and pET11cjoe (a kind gift from H. J. Khackmuss, Stuttgart University, Germany) that belong to incompatible groups and have distinct antibiotic resistance genes for selection and maintenance in E. coli strains.
2.4. Construction of Plasmids for Expression in E. coli Cells
The p11cjoe/barstar-EGFP plasmid contains barstar (the inhibitor of barnase) gene from Bacillus amyloliquefaciens and EGFP gene from Aequoria victoria fused in frame ORFs (linker sequence Gly-Thr-Gly). The barstar gene was amplified on pMT416 [23] as a template using bs_fw (5′-TTATTTCATATGAAAAAAGCAGTCATTAACGg-3′) and bs_rev (5′-ttggtacccggAGAAAGTATGATGGTGAT-3′) primers and cloned into the NdeI/KpnI sites of pET11cjoe plasmid to produce p11cjoe/barstar. The EGFP ORF was amplified on pEGFP (ClonTech Laboratories, Palo Alto, CA, USA) with the pair of primers EGFP_fw (5′-TAGGTACCGGTGTGAGCAAGGGCGAGGA-3′) and EGFP_rev (5’-ttgtcgacTTACTTGTACAGCTCGTCCATGc-3′), followed by cloning into KpnI/SalI sites of p11cjoe/barstar to yield p11cjoe/barstar-EGFP.
The plasmid p28/barnase-RLuc (linker sequence Pro-Glu-Phe-Gly) was constructed as follows. The ORF of Renilla luciferase (RLuc) was amplified using pET30a-RLuc as a template with the primers RLuc_fw (5′-ttgaattcggtACTTCGAAAGTTTATGATCC-3′) and RLuc_rev (5′-TTGTCGACTTATTGTTCATTTTTGAGAAC-3′) and cloned into EcoRI/SalI digested pAC28. The sequence encoding mature barnase protein was amplified with Bn_fw (5′-ttccatgggtGCACAGGTTATCAACAC-3′) and Bn_rev (5′-TTGAATTCCGGTCTGATTTTTGTAAAGGTC-3′) primers from pMT416 and cloned into NcoI/EcoRI digested pAC28-RLuc to give p28/barnase-RLuc. The plasmid constructs were controlled by sequencing.
2.5. Expression of the Genetic Constructs in E. coli
E. coli BL21(DE3) cells transformed with the recombinant plasmids were grown at 37 °C in 200 mL LB medium containing 100 µg/mL ampicillin in a shaking flask up to absorbance A590 of 0.5–0.6. After induction with IPTG (0.05 mM), the temperature was decreased to 20 °C and cells were grown for an additional period of 18 h.
2.6. Registration of Fluorescence and Bioluminescence Spectra
To register luminescence spectra of the cells a Varian Cary Eclipse spectrofluorometer (USA) was used. Cell suspension was diluted to OD590 = 1 with buffer (20 mM Tris-HCl, pH 7.6, 150 mM NaCl), and 20 µL of the suspension was mixed with 60 µL buffer in quartz microcuvette (3 × 3 × 5 mm). The fluorescence spectrum was measured within the wavelength range from 488 nm to 730 nm at 470 nm excitation and 10 nm slits. To measure bioluminescence spectra, 2 µL coelenterazine (1 mM in menthol) was added to cell suspension or cell lysate, and spectra were recorded within the 350–730 nm range with the excitation lamp turned off. The bioluminescence spectra were corrected for time-dependent change of bioluminescence intensity. Ten spectra were averaged, and the resultant one was corrected for the spectral sensitivity of the device [22].
3. Results
Generation and Identification of Fusion Proteins Barnase–RLuc and Barstar–EGFP
One of the main problems arising during the expression of recombinant fused genes is the correct folding of the resulting proteins. Our fusion proteins contain two protein modules: one is a photoprotein (luciferase or EGFP) and the other is an interacting protein (barnase or barstar); these are connected by a linker of three or four amino acids.
Table 1 presents the data on cell growth after transformation with corresponding plasmids.

Table 1.
E. coli cell growth after transformation with plasmids used.
One can see that the growth of cells transformed with the plasmid encoding barnase–luciferase is far from that without barnase, while after co-expression of plasmids encoding both fused proteins, the cell growth is normal. This is caused by the toxicity of barnase in the absence of its inhibitor, barstar [23], and hints at correct (functionally active) folding of barnase in the barnase–luciferase fusion, as well as barstar in the barstar–EGFP fusion.
The correct folding of photoproteins was monitored by their luminescence spectra. Figure 1 shows bioluminescence and fluorescence spectra of E. coli cells or cell lysates after the expression of the appropriate genes.
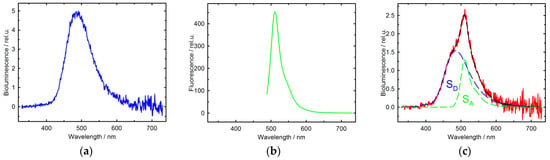
Figure 1.
Emission spectra of E. coli cells: Bioluminescence (a) and Fluorescence (λexs 470 nm) (b) spectra of the mixture of cell lysates after expression p11cjoe/RLuc and p11cjoe/barstar-EGFP; (c) The bioluminescence spectrum of E. coli cells after co-expression of p28/barnase-RLuc and p11cjoc/barstar-EGFP plasmids.
These spectra allow us to conclude the following:
First, the photoproteins (luciferase and EGFP) are correctly folded after the expression of their genes in E. coli cells, because they display their specific emission, except the fusion barnase–luciferase protein whose bioluminescence is immeasurable probably due to the toxicity of barnase in the absence of barstar (see Table 1).
Second, the bioluminescence spectrum of cells after co-expression of two plasmids encoding the fusion proteins barnase–luciferase and barstar–EGFP displays a pronounced EGFP emission, thereby unequivocally confirming the proximity of these proteins within living cells.
Third, luciferase from Renilla reniformis and the EGFP mutant of Aequoria victoria do not interact. This follows from the fact that after the expression of the luciferase gene and the barstar–EGFP fusion gene (Figure 1a), the bioluminescence spectrum of the cell lysate mixture does not show the GFP emission band. This guarantees that BRET observed in E. coli cells after co-expression of genes encoding the fusion proteins barnase–luciferase and barstar–EGFP (Figure 1c) results not from an interaction between photoproteins, but from that between barnase and barstar.
4. Discussion
One aspect of the present research needs to be discussed. It concerns the evaluation of the distance between the chromophores participating in the emission resonance energy transfer according to the Forster theory [26]. It looks promising to evaluate the distance between the chromophores of photoproteins fused with barnase and barstar within the complex, because the crystal structures of the barnase–barstar complex and photoproteins used are well established. The first step is the evaluation of the BRET efficiency. Unfortunately, there is a problem to use the known methods [26], which require the determination of the donor emission lifetime or intensity in the absence and presence of the acceptor. The difficulty is that it is impossible to compare in vivo the spectra of the donor (luciferase) in the absence and presence of the acceptor (EGFP), because we cannot determine the protein concentration and ensure equal conditions of registration of the spectra. Therefore, we propose that the efficiency of BRET (E) may be evaluated through decomposition of the bioluminescence spectrum (Figure 1c) into two components: the area of the donor, luciferase (SD), and the area of the acceptor, EGFP (SA), according to:
where QD is the quantum yield of the donor RLuc (0.053 [18]), and QA is the quantum yield of the acceptor EGFP (0.6 [27]).
E = (SA/QA)/(SD/QD + SA/QA),
The distance between chromophores of the donor and the acceptor (R) may be evaluated using the well-known Forster equation [26]:
where R0 is the Forster radius for 50% energy transfer between the definite donor–acceptor pair that may be evaluated from the known equation [26]:
where is the overlapping integral of bioluminescence spectra of the donor and absorption spectra of the acceptor (Figure 2a).
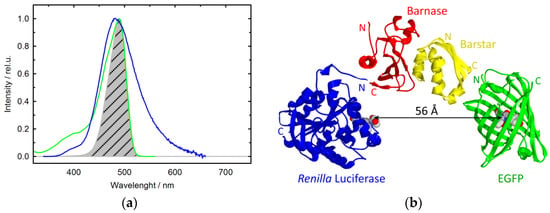
Figure 2.
Estimation of the distance between RLuc and EGFP chromophores within the complex of barnase–RLuc and barstar–EGFP: (a) Normalized spectra of RLuc bioluminescence (blue), EGFP absorption (green), and their overlapping integral (shaded); (b) Schematic presentation of the complex of barnase and barstar fused with photoproteins. The arrow indicates the evaluated distance between photoprotein chromophores (the crystal structures of the complex barnase–barstar, Renilla luciferase, and EGFP are taken from PDB files 1bgs, 2psj, and 2y0g, respectively, while the presentation was composed using SPDBViewer v4.1.0 software [28]).
Thus, the R0 value for the donor–acceptor pair RLuc-EGFP is estimated to be equal to 32 Å, while the average distance between their chromophores within the complex of barnase–RLuc with barstar–EGFP is estimated as 56 ± 2 Å. The error ± 2 Å does not signify the rigidity of the photoproteins orientation, but shows only the accuracy of the average distance determination by several experiments. The same distance may be realized at other orientations of the photoproteins, as far as allowed by the linkers. The basic parameters for these estimations are given in Table 2.

Table 2.
BRET parameters for the complex of barnase and barstar fused with RLuc and EGFP, respectively.
It should be emphasized that this distance is realistic for a complex of fusion proteins (Figure 2b).
Author Contributions
Conceptualization, V. K. and G.S.; Data curation, G.S.; Formal analysis, N.M.; Funding acquisition, G.S.; Investigation, V.M.; Methodology, V.M.; Project administration, N.M.; Resources, T.I.; Software, V.M.; Supervision, G.S.; Validation, V.M., V.K., and G. S.; Visualization, V.M.; Writing—original draft, Victor Marchenkov and Tanya Ivashina; Writing—review and editing, G.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 14-24-00157.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to E. Serebrova for the English correction.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Haddock, S.H.D.; Moline, M.A.; Case, J.F. Bioluminescence in the Sea. Annu. Rev. Mar. Sci. 2010, 2, 443–493. [Google Scholar] [CrossRef] [Green Version]
- Shimomura, O. Bioluminescence in the sea: Photoprotein systems. Symp. Soc. Exp. Biol. 1985, 39, 352–371. [Google Scholar]
- Ward, W.; Cormier, M. An energy transfer protein in coelenterate bioluminescence. Characterization of the Renilla green-fluorescent protein. J. Biol. Chem. 1979, 254, 781–788. [Google Scholar] [CrossRef]
- Gorokhovatsky, A.Y.; Marchenkov, V.V.; Rudenko, N.V.; Ivashina, T.V.; Ksenzenko, V.N.; Burkhardt, N.; Semisotnov, G.V.; Vinokurov, L.M.; Alakhov, Y.B. Fusion of Aequorea victoria GFP and aequorin provides their Ca2+-induced interaction that results in red shift of GFP absorption and efficient bioluminescence energy transfer. Biochem. Biophys. Res. Commun. 2004, 320, 703–711. [Google Scholar] [CrossRef]
- Wu, P.; Brand, L. Resonance Energy Transfer: Methods and Applications. Anal. Biochem. 1994, 218, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Morise, H.; Shimomura, O.; Johnson, F.H.; Winant, J. Intermolecular energy transfer in the bioluminescent system of Aequorea. Biochemistry 1974, 13, 2656–2662. [Google Scholar] [CrossRef]
- De, A.; Jasani, A.; Arora, R.; Gambhir, S.S. Evolution of BRET Biosensors from Live Cell to Tissue-Scale In vivo Imaging. Front. Endocrinol. 2013, 4, 131. [Google Scholar] [CrossRef] [Green Version]
- Dale, N.C.; Johnstone, E.K.M.; White, C.W.; Pfleger, K.D.G. NanoBRET: The Bright Future of Proximity-Based Assays. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef]
- Yeh, H.W.; Ai, H.-W. Development and Applications of Bioluminescent and Chemiluminescent Reporters and Biosensors. Annu. Rev. Anal. Chem. 2019, 12, 129–150. [Google Scholar] [CrossRef] [Green Version]
- Shimomura, O.; Johnson, F.H. Chemical nature of the light emitter in bioluminescence of aequorin. Tetrahedron Lett. 1973, 14, 2963–2966. [Google Scholar] [CrossRef]
- Xu, Y.; Piston, D.W.; Johnson, C.H. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. 1999, 96, 151–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audet, M.; Lagacé, M.; Silversides, D.W.; Bouvier, M. Protein-protein interactions monitored in cells from transgenic mice using bioluminescence resonance energy transfer. FASEB J. 2010, 24, 2829–2838. [Google Scholar] [CrossRef] [PubMed]
- Buckle, A.; Schreiber, G.; Fersht, A.R. Protein-protein recognition: Crystal structural analysis of a barnase-barstar complex at 2.0-.ANG. resolution. Biochemistry 1994, 33, 8878–8889. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Fersht, A.R. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry 1993, 32, 5145–5150. [Google Scholar] [CrossRef]
- Shilova, O.; Kotelnikova, P.; Proshkina, G.; Shramova, E.; Deyev, S. Barnase-Barstar Pair: Contemporary Application in Cancer Research and Nanotechnology. Molecules 2021, 26, 6785. [Google Scholar] [CrossRef]
- Brown, N.E.; Blumer, J.B.; Hepler, J.R. Bioluminescence resonance energy transfer to detect protein-protein interactions in live cells. In Protein-Protein Interactions; Humana Press: New York, NY, USA, 2015; pp. 457–465. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.W.; Samanta, A.; Díaz, S.A.; Buckhout-White, S.; Walper, S.A.; Goldman, E.R.; Medintz, I.L. Dendrimeric DNA Nanostructures as Scaffolds for Efficient Bidirectional BRET–FRET Cascades. Adv. Opt. Mater. 2017, 5, 1700181. [Google Scholar] [CrossRef]
- Loening, A.M.; Fenn, T.D.; Wu, A.M.; Gambhir, S.S. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006, 19, 391–400. [Google Scholar] [CrossRef] [Green Version]
- Issad, T.; Boute, N.; Boubekeur, S.; Lacasa, D. Interaction of PTPB with the insulin receptor precursor during its biosynthesis in the endoplasmic reticulum. Biochimie 2005, 87, 111–116. [Google Scholar] [CrossRef]
- Kamal, M.; Marquez, M.; Vauthier, V.; Leloire, A.; Froguel, P.; Jockers, R.; Couturier, C. Improved donor/acceptor BRET couples for monitoring β-arrestin recruitment to G protein-coupled receptors. Biotechnol. J. 2009, 4, 1337–1344. [Google Scholar] [CrossRef] [Green Version]
- Gomes, I.; Filipovska, J.; Jordan, B.A.; Devi, L.A. Oligomerization of opioid receptors. Methods 2002, 27, 358–365. [Google Scholar] [CrossRef]
- Gorokhovatsky, A.Y.; Rudenko, N.V.; Marchenkov, V.V.; Skosyrev, V.S.; A Arzhanov, M.; Burkhardt, N.; Zakharov, M.V.; Semisotnov, G.; Vinokurov, L.M.; Alakhov, Y.B. Homogeneous assay for biotin based on Aequorea victoria bioluminescence resonance energy transfer system. Anal. Biochem. 2003, 313, 68–75. [Google Scholar] [CrossRef]
- Hartley, R.W. Barnase and barstar: Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J. Mol. Biol. 1988, 202, 913–915. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.J.; Russell, D.D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 0-87969-577-3. [Google Scholar]
- Kholod, N.; Mustelin, T. Novel Vectors for Co-Expression of Two Proteins in E. coli. BioTechniques 2001, 31, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Boston, MA, USA, 2006. [Google Scholar]
- Patterson, G.; Knobel, S.; Sharif, W.; Kain, S.; Piston, D. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997, 73, 2782–2790. [Google Scholar] [CrossRef] [Green Version]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).