Molecular Epidemiology of SARS-CoV-2 in Northern Greece from the Index Case up to Early 2025 Using Nanopore Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Library Preparation/Sequencing Using MinION Mk1C and GridION
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Worldometer-Coronavirus. Available online: https://www.worldometers.info/coronavirus (accessed on 2 September 2025).
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar]
- Lippi, G.; Mattiuzzi, C.; Henry, B.M. Updated picture of SARS-CoV-2 variants and mutations. Diagnosis 2021, 9, 11–17. [Google Scholar]
- Gong, W.; Parkkila, S.; Wu, X.; Aspatwar, A. SARS-CoV-2 variants and COVID-19 vaccines: Current challenges and future strategies. Int. Rev. Immunol. 2023, 42, 393–414. [Google Scholar] [CrossRef]
- Nakagawa, S.; Miyazawa, T. Genome evolution of SARS-CoV-2 and its virological characteristics. Inflamm. Regen. 2020, 40, 17. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [PubMed]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Fawaz, M.A.M.; Aisha, A. A comparative overview of SARS-CoV-2 and its variants of concern. Infez. Med. 2022, 30, 328–343. [Google Scholar] [PubMed]
- Scovino, A.M.; Dahab, E.C.; Vieira, G.F.; Freire-de-Lima, L.; Freire-de-Lima, C.G.; Morrot, A. SARS-CoV-2’s Variants of Concern: A brief characterization. Front. Immunol. 2022, 13, 834098. [Google Scholar] [CrossRef]
- Chen, K.W.K.; Huang, D.T.N.; Huang, L.M. SARS-CoV-2 variants–Evolution, spike protein, and vaccines. Biomed. J. 2022, 45, 573–579. [Google Scholar] [CrossRef]
- Stefanelli, P.; Faggioni, G.; Lo Presti, A.; Fiore, S.; Marchi, A.; Benedetti, E.; Fabiani, M.; Anselmo, A.; Ciammaruconi, A.; Fortunato, A.; et al. Whole Genome and Phylogenetic Analysis of Two SARS-CoV-2 Strains Isolated in Italy in January and February 2020. Euro Surveill. 2020, 25, 2000603. [Google Scholar]
- Maltezou, H.C.; Papadima, K.; Gkolfinopoulou, K.; Ferentinos, G.; Mouratidou, E.; Andreopoulou, A.; Pavli, A.; Magaziotou, I.; Georgakopoulou, T.; Mellou, K.; et al. Coronavirus disease 2019 pandemic in Greece, February 26–May 3, 2020: The first wave. Travel Med. Infect. Dis. 2021, 41, 102051. [Google Scholar] [CrossRef]
- Di Giallonardo, F.; Duchene, S.; Puglia, I.; Curini, V.; Profeta, F.; Cammà, C.; Marcacci, M.; Calistri, P.; Holmes, E.C.; Lorusso, A. Genomic Epidemiology of the First Wave of SARS-CoV-2 in Italy. Viruses 2020, 12, 1438. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Hodcroft, E.B.; Zuber, M.; Nadeau, S.; Crawford, K.H.D.; Bloom, J.D.; Veesler, D.; Vaughan, T.G.; Comas, I.; Candelas, F.G.; Stadler, T.; et al. Spread of a SARS-CoV-2 Variant through Europe in the Summer of 2020. Nature 2021, 595, 707–712. [Google Scholar]
- Brejová, B.; Boršová, K.; Hodorová, V.; Čabanová, V.; Reizigová, L.; Paul, E.D.; Čekan, P.; Klempa, B.; Nosek, J.; Vinař, T. A SARS-CoV-2 Mutant from B.1.258 Lineage with ∆H69/∆V70 Deletion in the Spike Protein Circulating in Central Europe in the Fall 2020. Virus Genes 2021, 57, 556–560. [Google Scholar]
- Sakaloglou, P.; Bozidis, P.; Kourou, K.; Kostoulas, C.; Gouni, A.; Tsaousi, E.; Koumpouli, D.; Argyropoulou, S.; Oikonomidis, P.; Peponi, H.; et al. Genomic and Epidemiological Surveillance of SARS-CoV-2 Epidemic in Northwestern Greece. Acta Microbiol. Hell. 2024, 69, 285–294. [Google Scholar]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [PubMed]
- Calistri, P.; Amato, L.; Puglia, I.; Cito, F.; Di Domenico, M.; Danzetta, M.L.; Caporale, M.; Di Gennaro, A.; Ancora, M.; Curini, V.; et al. Infection Sustained by Lineage B.1.1.7 of SARS-CoV-2 Is Characterized by Longer Persistence and Higher Viral RNA Loads in Nasopharyngeal Swabs. Int. J. Infect. Dis. 2021, 105, 753–755. [Google Scholar] [PubMed]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased Transmissibility and Global Spread of SARS-CoV-2 Variants of Concern as at June 2021. Euro Surveill. 2021, 26, 2100509. [Google Scholar] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Kant, R.; Nguyen, P.T.; Blomqvist, S.; Erdin, M.; Alburkat, H.; Suvanto, M.; Zakham, F.; Salminen, V.; Olander, V.; Paloniemi, M.; et al. Incidence Trends for SARS-CoV-2 Alpha and Beta Variants, Finland, Spring 2021. Emerg. Infect. Dis. 2021, 27, 3137–3141. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 Delta (B.1.617.2) compared with Alpha (B.1.1.7): A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [PubMed]
- Meletis, G.; Tychala, A.; Ntritsos, G.; Verrou, E.; Savvidou, F.; Dermitzakis, I.; Chatzidimitriou, A.; Gkeka, I.; Fyntanidou, B.; Gkarmiri, S.; et al. Variant-Related Differences in Laboratory Biomarkers among Patients Affected with Alpha, Delta and Omicron: A Retrospective Whole Viral Genome Sequencing and Hospital-Setting Cohort Study. Biomedicines 2023, 11, 1143. [Google Scholar]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Lessells, R.J.; Giandhari, J.; Wolter, N.; Everatt, J.; Rambaut, A.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in Southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalisation and death associated with Omicron (B.1.1.529) and Delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar]
- Català, M.; Coma, E.; Alonso, C.; Andrés, E.; Blanco, I.; Antón, A.; Bordoy, M.; Cardona, G.; Fina, L.; Martró, E.; et al. Transmissibility, Hospitalization, and Intensive Care Admissions Due to Omicron Compared to Delta Variants of SARS-CoV-2 in Catalonia: A Cohort Study and Ecological Analysis. Front. Public Health 2022, 10, 961030. [Google Scholar]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 615, 572–582. [Google Scholar] [CrossRef]
- VanInsberghe, D.; Neish, A.S.; Lowen, A.C.; Koelle, K. Recombinant SARS-CoV-2 genomes circulated at low levels over the first year of the pandemic. Virus Evol. 2021, 7, veab059. [Google Scholar] [CrossRef]
- World Health Organization. Statement on EG.5 Initial Risk Evaluation (IRE); WHO: Geneva, Switzerland, 2023; Available online: https://www.who.int/docs/default-source/coronaviruse/09082023eg.5_ire_final.pdf (accessed on 2 October 2025).
- World Health Organization. COVID-19—Global Situation; WHO: Geneva, Switzerland, 2025; Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2025-DON572 (accessed on 2 October 2025).
- Shanmugaraj, B. Emergence of SARS-CoV-2 variants KP.2 and KP.3 sparks concerns: What should we do? Health Care Sci. 2024, 3, 211–214. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef]
- Tyler, A.D.; Mataseje, L.; Urfano, C.J.; Schmidt, L.; Antonation, K.S.; Mulvey, M.R.; Corbett, C.R. Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep. 2018, 8, 10931. [Google Scholar] [CrossRef]
- Charre, C.; Ginevra, C.; Sabatier, M.; Regue, H.; Destras, G.; Brun, S.; Burfin, G.; Scholtes, C.; Morfin, F.; Valette, M.; et al. Evaluation of NGS-based approaches for SARS-CoV-2 whole genome characterization. Virus. Evol. 2020, 6, veaa075. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION sequencing and genome assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G.; Tychala, A.; Gkeka, I.; Gkotzia, A.; Triantafyllou, A.; Pappa, S.; Exindari, M.; Gioula, G.; Papa, A.; Skoura, L. Clinical Performance Evaluation of the NeuMoDx Flu A-B/RSV/SARS-CoV-2 Vantage Assay. Diagnostics 2022, 12, 3201. [Google Scholar] [CrossRef] [PubMed]
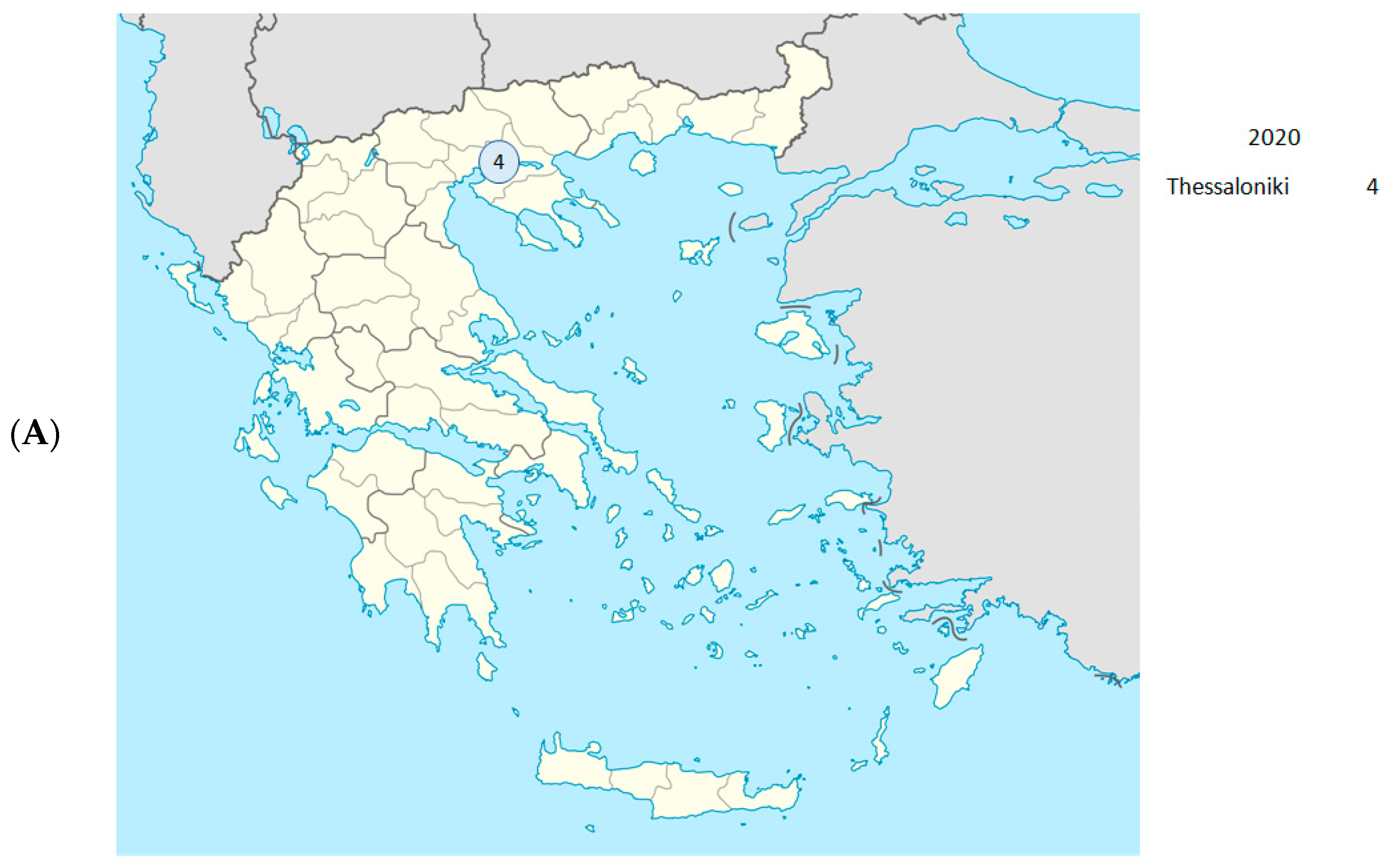
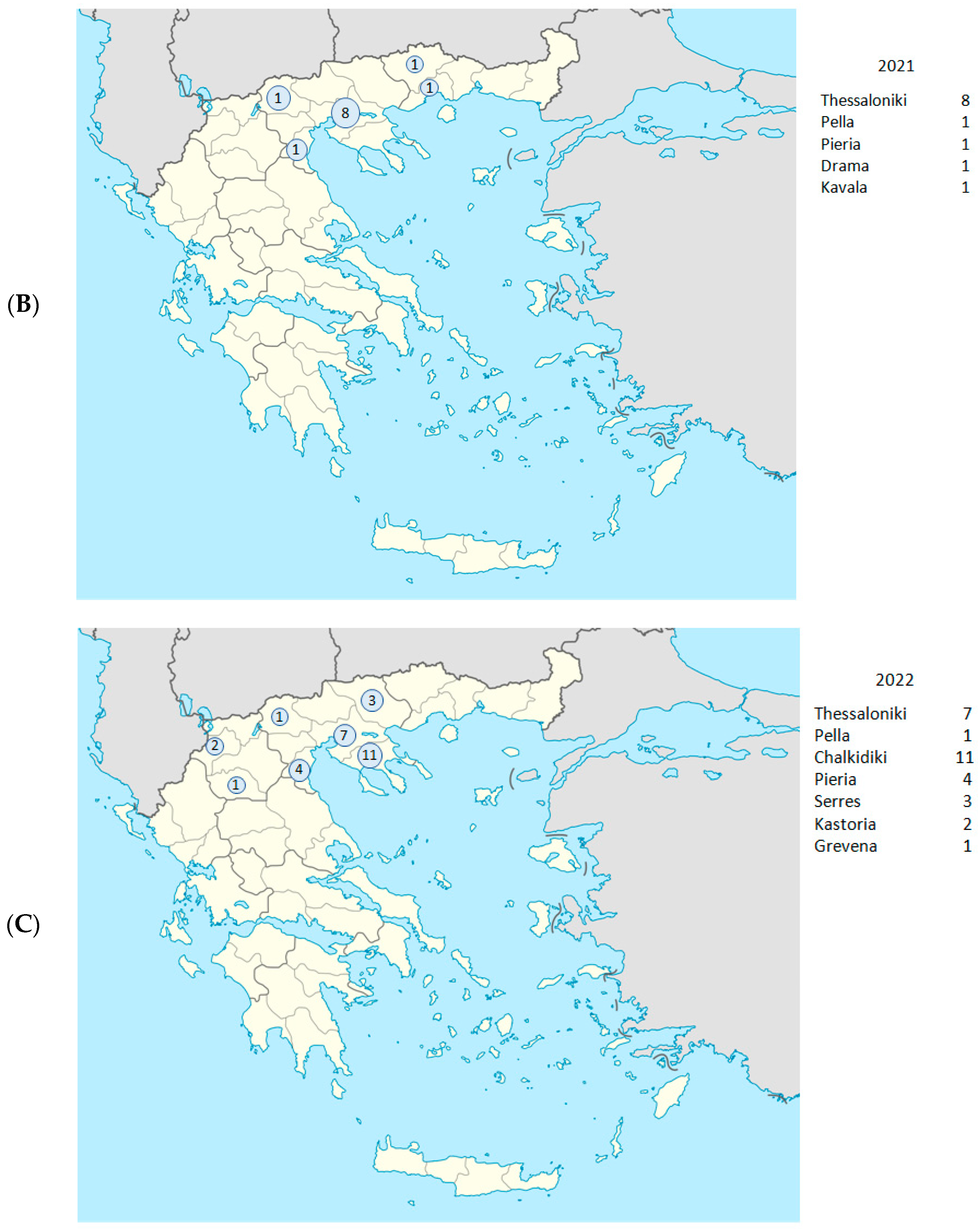
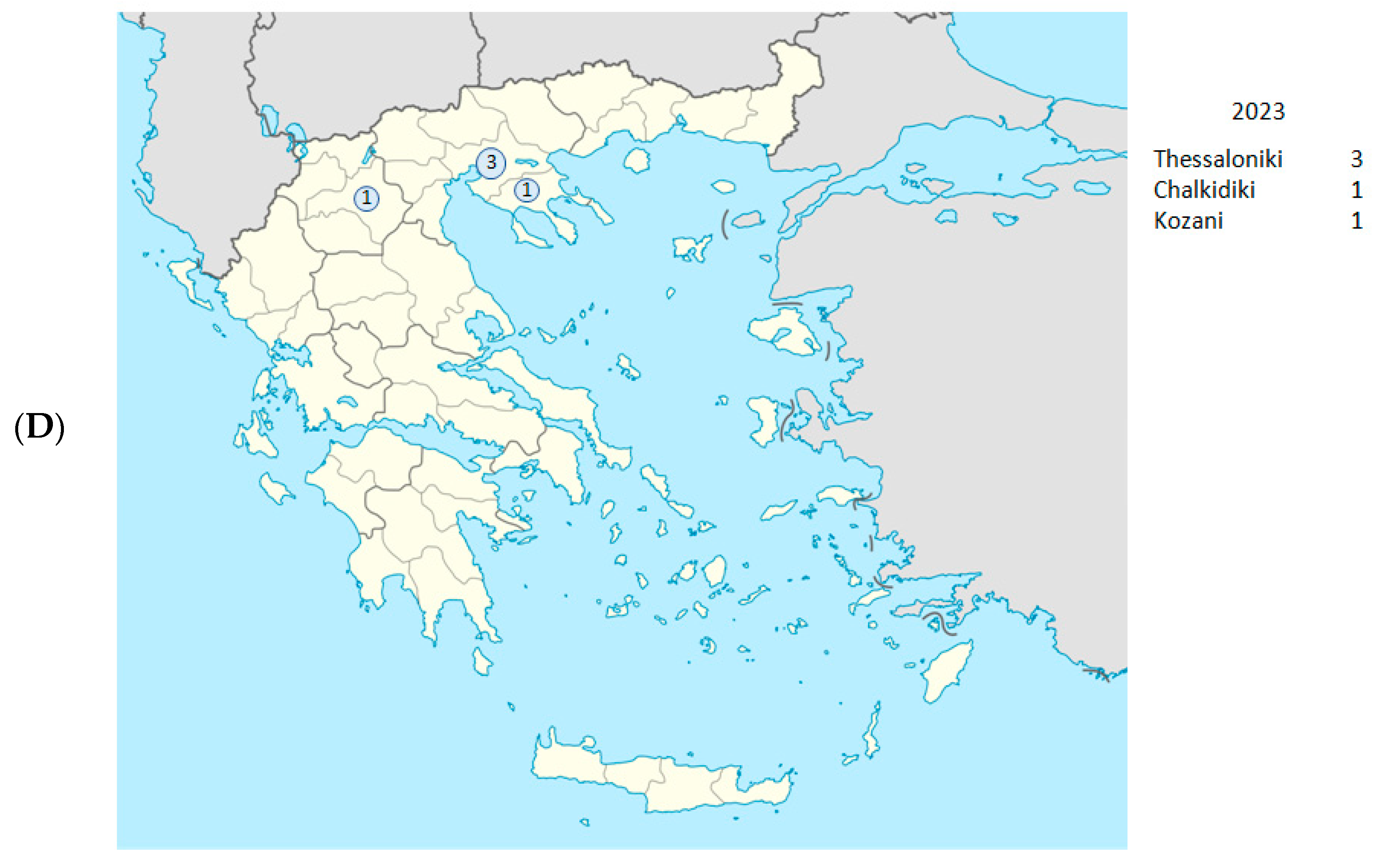
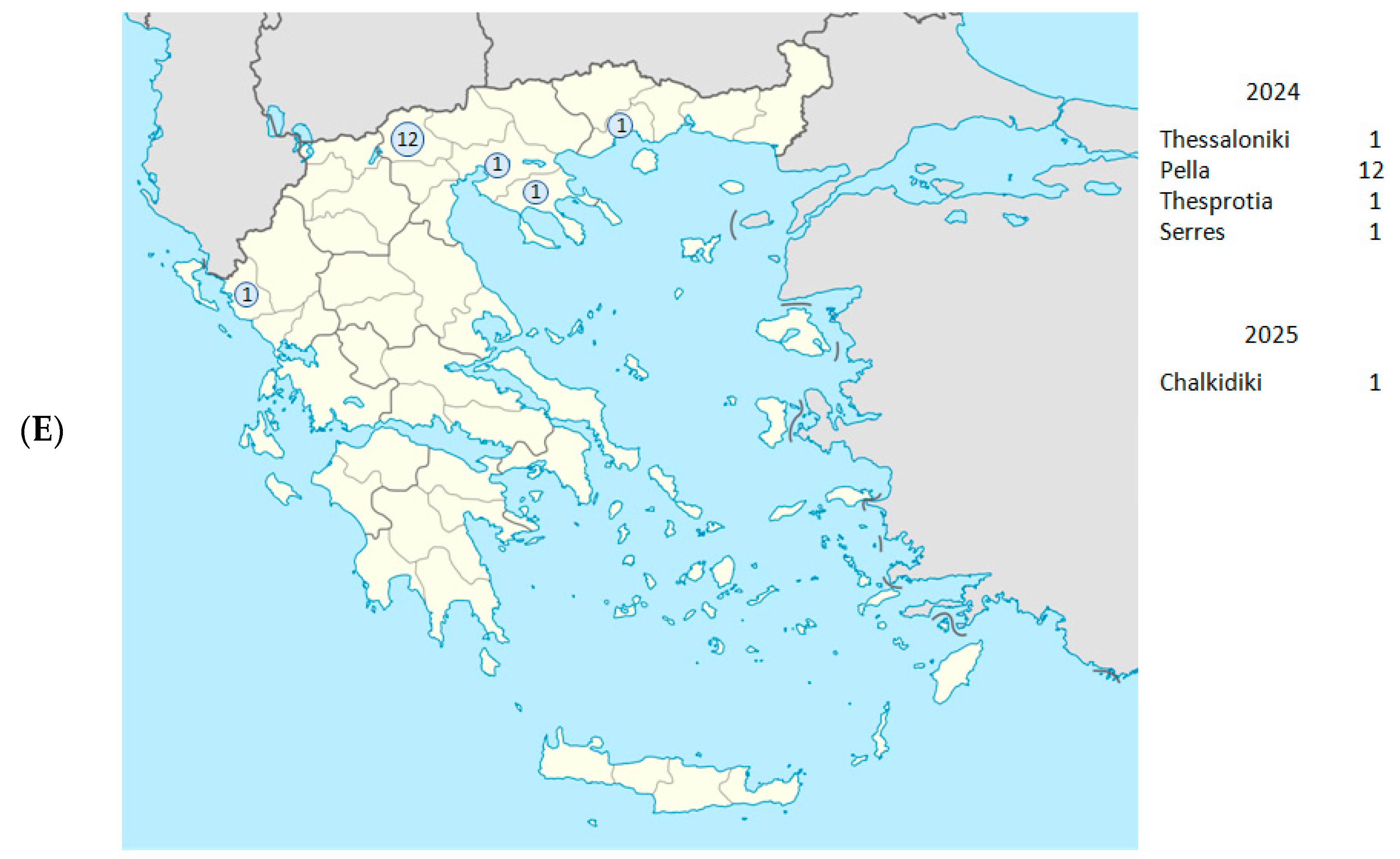

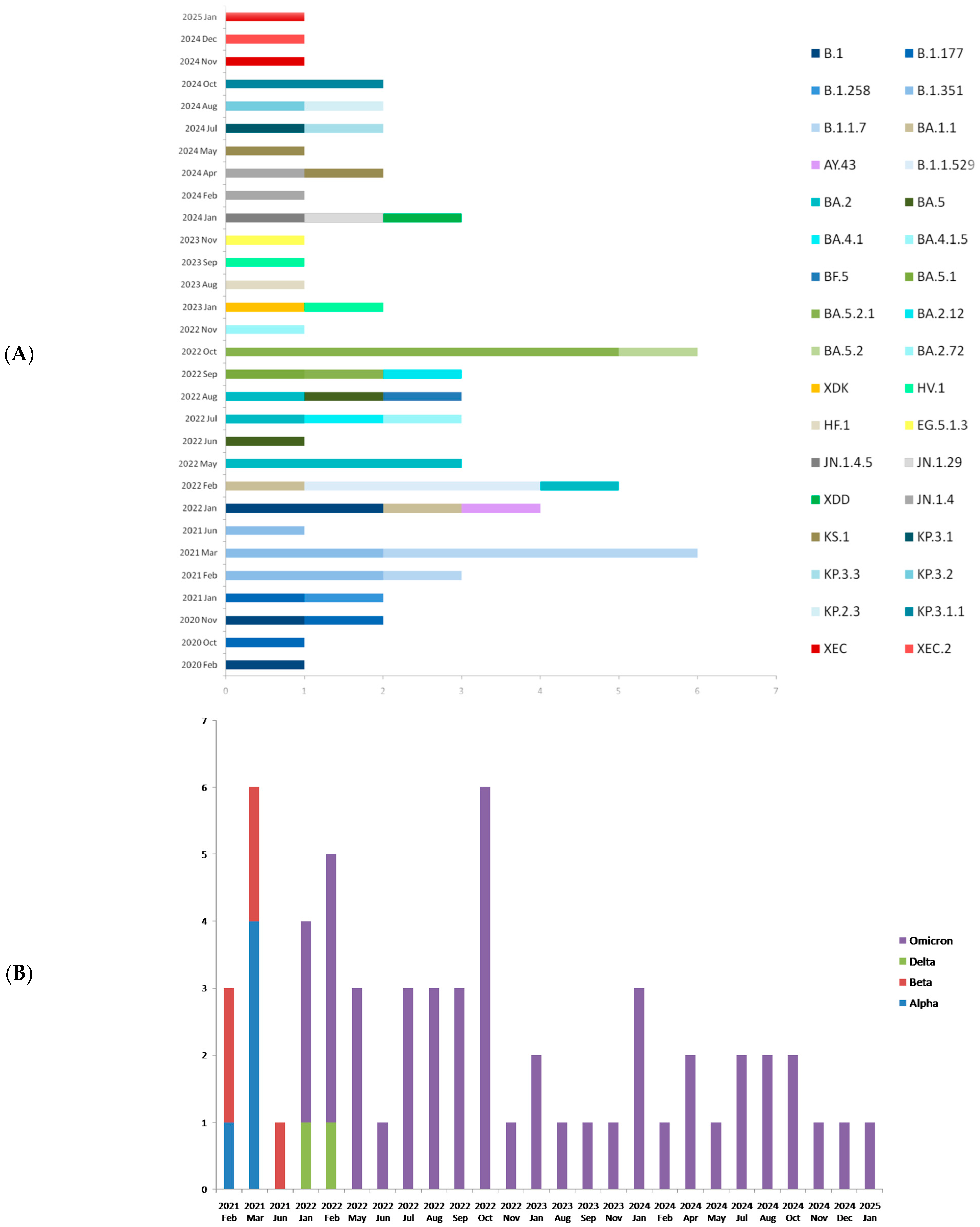
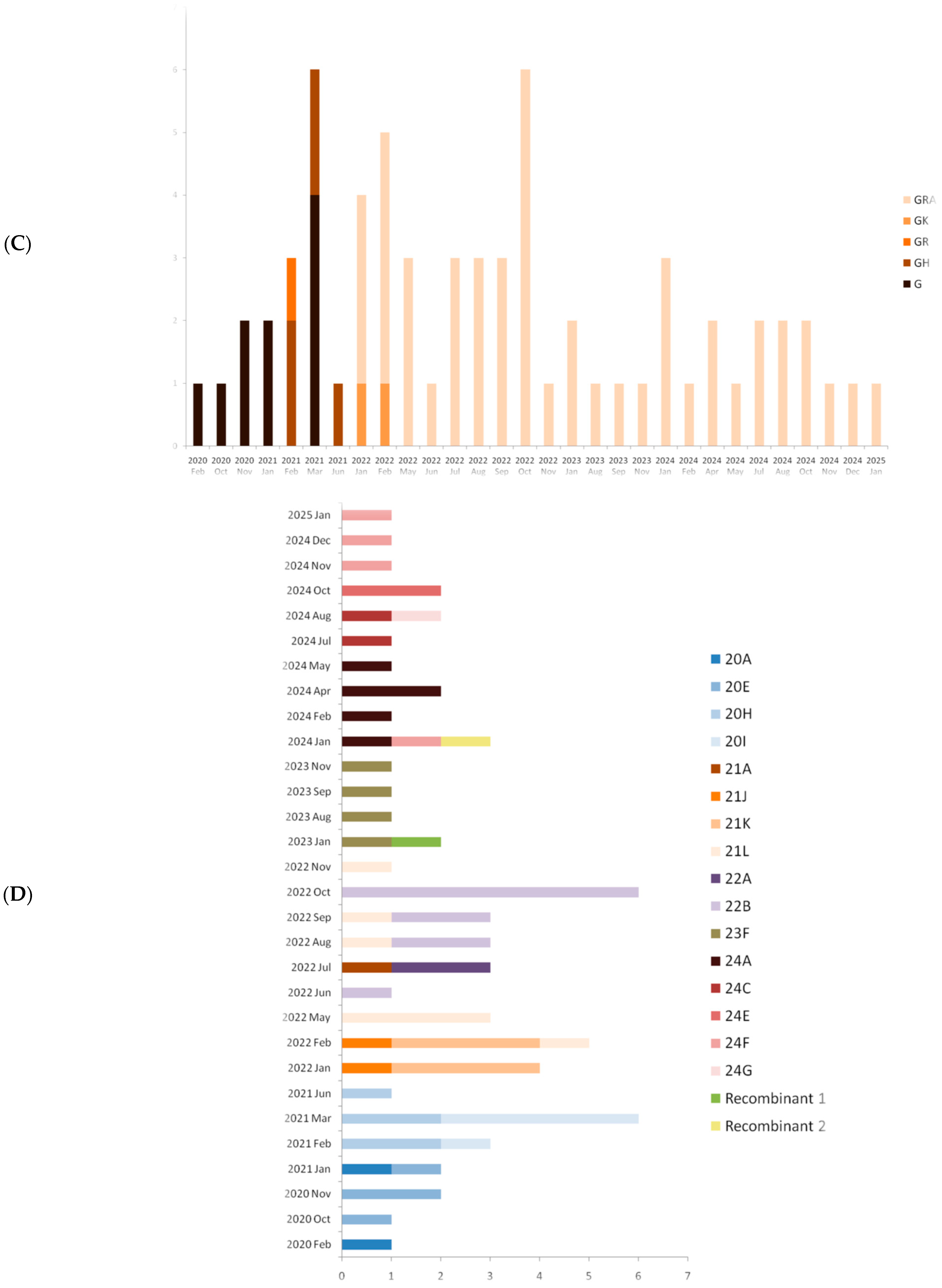
| Date (Day/ Month/Year) | Year- Week | Sample ID | Regional Unit of Residence | Hospital | PANGO Lineage | WHO Label | GISAID Clade | NextstrainClade |
|---|---|---|---|---|---|---|---|---|
| 29 February 2020 | 2020-W09 | 141 | Thessaloniki | AHEPA | B.1 | - | G | 20A |
| 29 October 2020 | 2020-W44 | Β10880 | Thessaloniki | Genimmatas | B.1.177 | - | G | 20E |
| 23 November 2020 | 2020-W48 | Γ4444 | Thessaloniki | Genimmatas | B.1 | - | G | 20E |
| 22 November 2020 | 2020-W47 | Γ4452 | Thessaloniki | Genimmatas | B.1.177 | - | G | 20E |
| 28 January 2021 | 2021-W04 | Δ4991 | Thessaloniki | Agios Dimitrios | B.1.177 | - | G | 20E |
| 28 January 2021 | 2021-W04 | Δ5147 | Kavala | Kavala | B.1.258 | - | G | 20A |
| 19 February 2021 | 2021-W07 | Δ8410 | Thessaloniki | Ippokrateio | B.1.351 | Beta | GH | 20H |
| 20 February 2021 | 2021-W07 | Δ8608 | Thessaloniki | Ippokrateio | B.1.351 | Beta | GH | 20H |
| 27 February 2021 | 2021-W08 | Δ9766 | Thessaloniki | Ippokrateio | B.1.1.7 | Alpha | GR | 20I |
| 1 March 2021 | 2021-W07 | Pg61 | Thessaloniki | Papageorgiou | B.1.1.7 | Alpha | G | 20I |
| 1 March 2021 | 2021-W09 | E109 | Thessaloniki | Ippokrateio | B.1.351 | Beta | GH | 20H |
| 5 March 2021 | 2021-W09 | E926 | Pieria | Katerini | B.1.351 | Beta | GH | 20H |
| 12 March 2021 | 2021-W10 | E1776 | Drama | Drama | B.1.1.7 | Alpha | G | 20I |
| 31 March 2021 | 2021-W13 | E4748 | Pella | Giannitsa | B.1.1.7 | Alpha | G | 20I |
| 17 March 2021 | 2021-W11 | Pg91 | Thessaloniki | Papageorgiou | B.1.1.7 | Alpha | G | 20I |
| 10 June 2021 | 2021-W23 | Ip21 | Thessaloniki | Ippokrateio | B.1.351 | Beta | GH | 20H |
| 14 January 2022 | 2022-W02 | O5490 | Grevena | Grevena | BA.1.1 | Omicron | GRA | 21K |
| 14 January 2022 | 2022-W02 | O5475 | Pieria | Katerini | BA.1 | Omicron | GRA | 21K |
| 15 January 2022 | 2022-W02 | O5476 | Pieria | Katerini | BA.1 | Omicron | GRA | 21K |
| 21 January 2022 | 2022-W03 | O6666 | Pieria | Katerini | AY.43 | Delta | GK | 21J |
| 1 February 2022 | 2022-W05 | Π322 | Thessaloniki | AHEPA | AY.43 | Delta | GK | 21J |
| 23 February 2022 | 2022-W08 | Π2997 | Thessaloniki | Papageorgiou | B.1.1.529 | Omicron | GRA | 21K |
| 23 February 2022 | 2022-W08 | Π3004 | Serres | Serres | B.1.1.529 | Omicron | GRA | 21K |
| 24 February 2022 | 2022-W08 | Π3005 | Kastoria | Kastoria | B.1.1.529 | Omicron | GRA | 21K |
| 28 February 2022 | 2022-W09 | Π3147 | Chalkidiki | Poligiros | BA.2 | Omicron | GRA | 21L |
| 18 May 2022 | 2022-W16 | T269 | Chalkidiki | Poligiros | BA.2 | Omicron | GRA | 21L |
| 23 May 2022 | 2022-W16 | Τ372 | Kastoria | Kastoria | BA.2 | Omicron | GRA | 21L |
| 26 May 2022 | 2022-W17 | T374 | Chalkidiki | Poligiros | BA.2 | Omicron | GRA | 21L |
| 31 May 2022 | 2022-W17 | Υ4 | Pieria | Katerini | BA.5 | Omicron | GRA | 22B |
| 31 May 2022 | 2022-W17 | Υ34 | Serres | Serres | BA.4.1 | Omicron | GRA | 22A |
| 12 July 2022 | 2022-W28 | Φ216 | Thessaloniki | Ippokrateio | BA.4.1.5 | Omicron | GRA | 22A |
| 14 July 2022 | 2022-W28 | Φ437 | Thessaloniki | Papanikolaou | BA.2 | Omicron | GRA | 21A |
| 21 August 2022 | 2022-W33 | Υ220 | Chalkidiki | Poligiros | BA.5 | Omicron | GRA | 22B |
| 1 August 2022 | 2022-W31 | Χ115 | Chalkidiki | Poligiros | BA.2 | Omicron | GRA | 21L |
| 1 August 2022 | 2022-W31 | Χ148 | Pella | Edessa | BF.5 | Omicron | GRA | 22B |
| 5 September 2022 | 2022-W36 | Ψ68 | Serres | Serres | BA.5.1 | Omicron | GRA | 22B |
| 2 September 2022 | 2022-W35 | Ψ60 | Chalikidiki | Poligiros | BA.5.2.1 | Omicron | GRA | 22B |
| 9 September 2022 | 2022-W36 | Ψ157 | Thessaloniki | Papanikolaou | BA.2.12 | Omicron | GRA | 21L |
| 6 October 2022 | 2022-W40 | Ω208 | Thessaloniki | Papanikolaou | BA.5.2.1 | Omicron | GRA | 22B |
| 6 October 2022 | 2022-W40 | Ω210 | Thessaloniki | Papanikolaou | BA.5.2.1 | Omicron | GRA | 22B |
| 26 October 2022 | 2022-W43 | Ω426 | Chalkidiki | Poligiros | BA.5.2 | Omicron | GRA | 22B |
| 23 October 2022 | 2022-W42 | Ω375 | Chalkidiki | Poligiros | BA.5.2.1 | Omicron | GRA | 22B |
| 28 October 2022 | 2022-W43 | Ω447 | Chalkidiki | Poligiros | BA.5.2.1 | Omicron | GRA | 22B |
| 26 October 2022 | 2022-W43 | Ω410 | Chalkidiki | Poligiros | BA.5.2.1 | Omicron | GRA | 22B |
| 1 November 2022 | 2022-W44 | 1A-10 | Chalkidiki | Poligiros | BA.2.72 | Omicron | GRA | 21L |
| 12 January 2023 | 2023-W02 | S1 | Thessaloniki | Ippokrateio | XDK | Omicron | GRA | Recombinant 1 |
| 14 January 2023 | 2023-W02 | D1 | Thessaloniki | Ippokrateio | HV.1 | Omicron | GRA | 23F |
| 3 August 2023 | 2023-W31 | C4172 | Kozani | Kozani | HF.1 | Omicron | GRA | 23F |
| 13 September 2023 | 2023-W37 | 1Λ-10 | Chalkidiki | Poligiros | HV.1 | Omicron | GRA | 23F |
| 17 November 2023 | 2023-W46 | C5066 | Thessaloniki | Ippokrateio | EG.5.1.3 | Omicron | GRA | 23F |
| 22 January 2024 | 2024-W04 | C5660 | Thesprotia | Filiaton | JN.1.4.5 | Omicron | GRA | 24F |
| 5 January 2024 | 2024-W01 | C5495 | Pella | Edessa | JN.1.29 | Omicron | GRA | 24A |
| 21 January 2024 | 2024-W51 | C5422 | Serres | Serres | XDD | Omicron | GRA | Recombinant 2 |
| 1 February 2024 | 2024-W05 | 1Λ353 | Pella | Giannitsa | JN.1.4 | Omicron | GRA | 24A |
| 8 April 2024 | 2024-W06 | 1Λ554 | Pella | Giannitsa | JN.1.4 | Omicron | GRA | 24A |
| 21 April 2024 | 2024-W16 | 1Λ593 | Pella | Giannitsa | KS.1 | Omicron | GRA | 24A |
| 2 May 2024 | 2024-W18 | 1Λ638 | Pella | Giannitsa | KS.1 | Omicron | GRA | 24A |
| 14 July 2024 | 2024-W28 | 1Λ761 | Pella | Giannitsa | KP.3.1 | Omicron | GRA | 24C |
| 29 July 2024 | 2024-W31 | 1Λ786 | Pella | Giannitsa | KP.3.3 | Omicron | GRA | 24C |
| 6 August 2024 | 2024-W32 | C5926 | Pella | Edessa | KP.3.2 | Omicron | GRA | 24C |
| 6 August 2024 | 2024-W32 | C5927 | Pella | Edessa | KP.2.3 | Omicron | GRA | 24G |
| 7 October 2024 | 2024-W41 | 1Λ867 | Pella | Giannitsa | KP.3.1.1 | Omicron | GRA | 24E |
| 5 October 2024 | 2024-W40 | C6016 | Thessaloniki | Ippokrateio | KP.3.1.1 | Omicron | GRA | 24E |
| 21 November 2024 | 2024-W47 | 1Λ907 | Pella | Giannitsa | XEC | Omicron | GRA | 24F |
| 27 November 2024 | 2024-W48 | C6098 | Pella | Edessa | XEC.2 | Omicron | GRA | 24F |
| 7 January 2025 | 2025-W02 | C6183 | Chalkidiki | Poligiros | XEC | Omicron | GRA | 24F |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meletis, G.; Pappa, S.; Gioula, G.; Exindari, M.; Christoforidi, M.; Papa, A. Molecular Epidemiology of SARS-CoV-2 in Northern Greece from the Index Case up to Early 2025 Using Nanopore Sequencing. Epidemiologia 2025, 6, 78. https://doi.org/10.3390/epidemiologia6040078
Meletis G, Pappa S, Gioula G, Exindari M, Christoforidi M, Papa A. Molecular Epidemiology of SARS-CoV-2 in Northern Greece from the Index Case up to Early 2025 Using Nanopore Sequencing. Epidemiologia. 2025; 6(4):78. https://doi.org/10.3390/epidemiologia6040078
Chicago/Turabian StyleMeletis, Georgios, Styliani Pappa, Georgia Gioula, Maria Exindari, Maria Christoforidi, and Anna Papa. 2025. "Molecular Epidemiology of SARS-CoV-2 in Northern Greece from the Index Case up to Early 2025 Using Nanopore Sequencing" Epidemiologia 6, no. 4: 78. https://doi.org/10.3390/epidemiologia6040078
APA StyleMeletis, G., Pappa, S., Gioula, G., Exindari, M., Christoforidi, M., & Papa, A. (2025). Molecular Epidemiology of SARS-CoV-2 in Northern Greece from the Index Case up to Early 2025 Using Nanopore Sequencing. Epidemiologia, 6(4), 78. https://doi.org/10.3390/epidemiologia6040078








