Genomic Surveillance of SARS-CoV-2: Data Analysis and Assessment of Tunisian Strategy from January 2021 to February 2022
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Study Population
2.3. Genomic Surveillance System Design
2.4. Audit Standards of the Tunisian Sequencing Strategy for SARS-CoV-2
2.5. Data Collection
2.6. Representativeness and Timeliness Evaluation
2.7. Ethical Approval
2.8. Analysis
2.8.1. Analysis of SARS-CoV-2 Genomic Surveillance Data
2.8.2. Evaluation of the National Sequencing Strategy
- -
- If there is total lack of required information or activities, the level of compliance is considered as “not met”;
- -
- If required information or activities are partially respected, the level of compliance is considered as “partially met”;
- -
- If we disposed of all the information needed and the activity was well realized, the level of compliance is considered as “fully met”.
3. Results
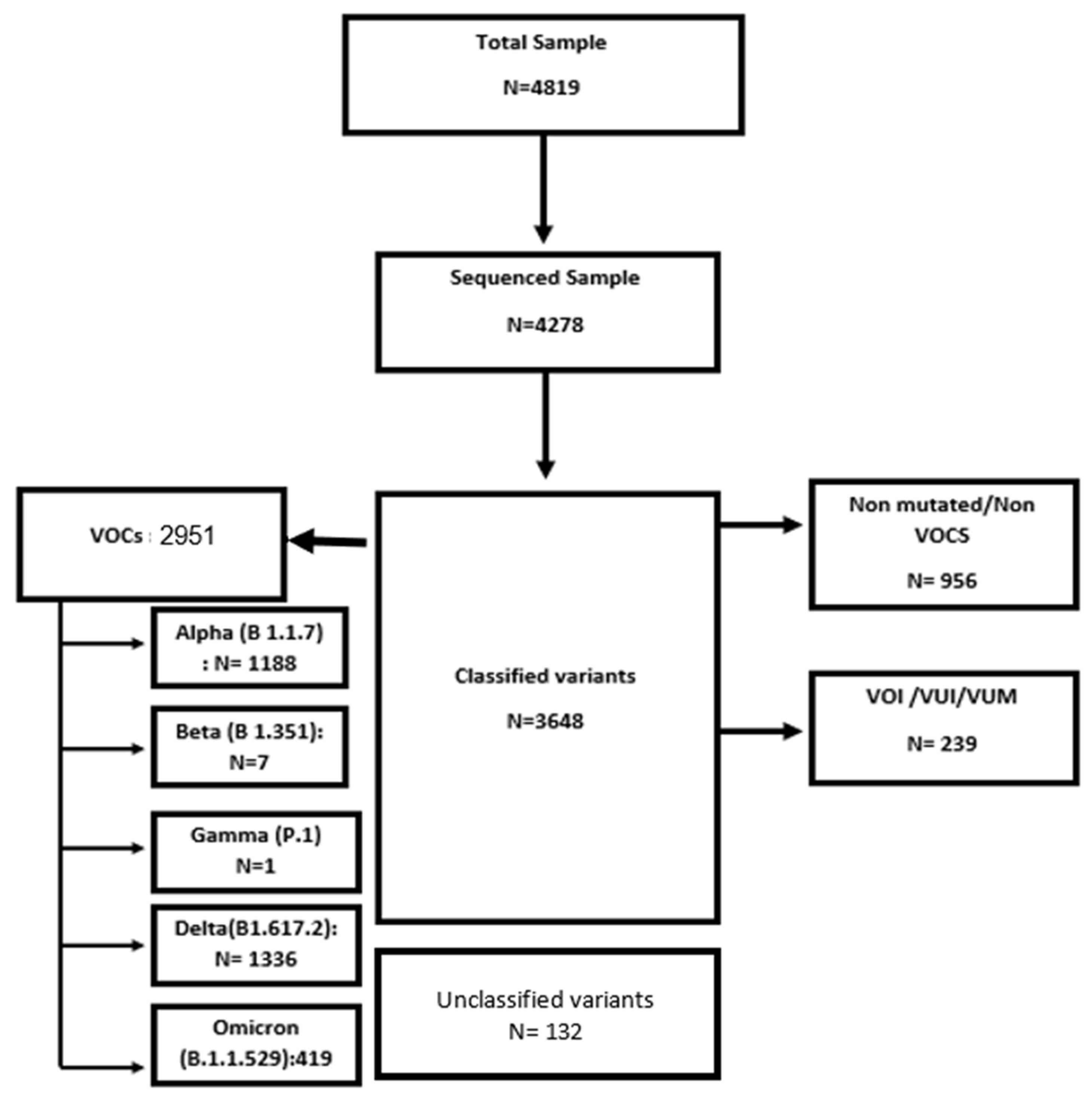
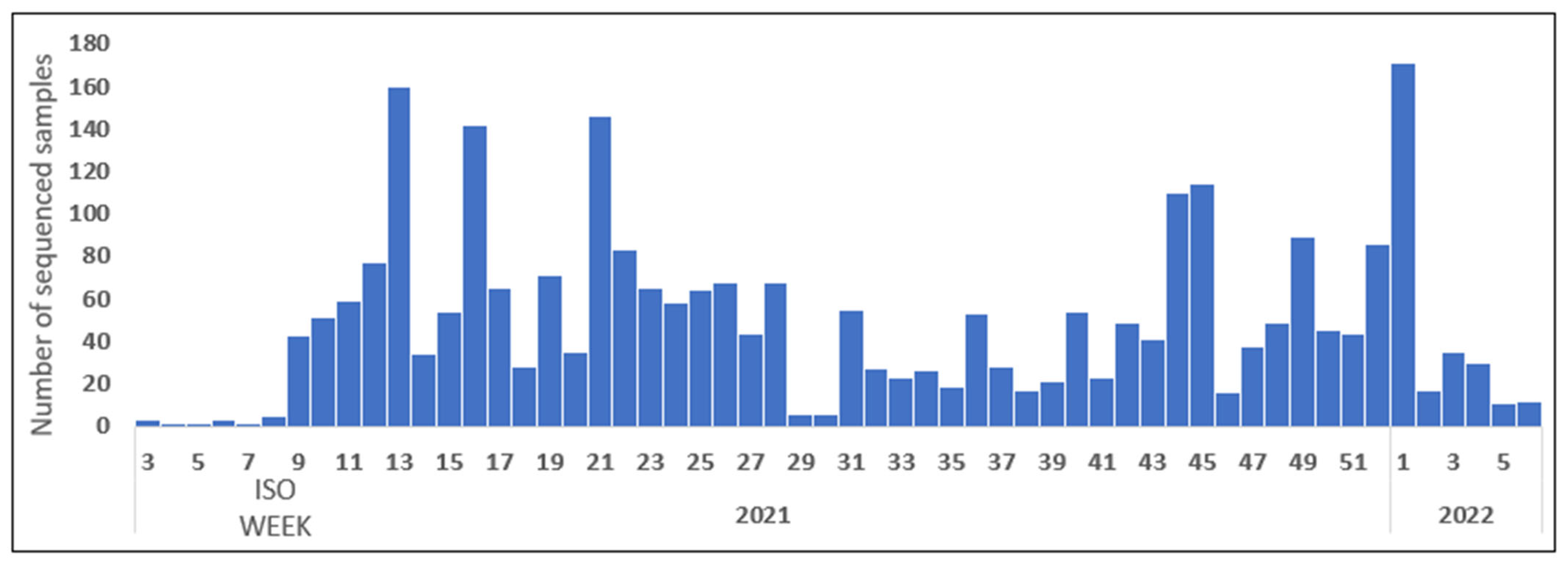
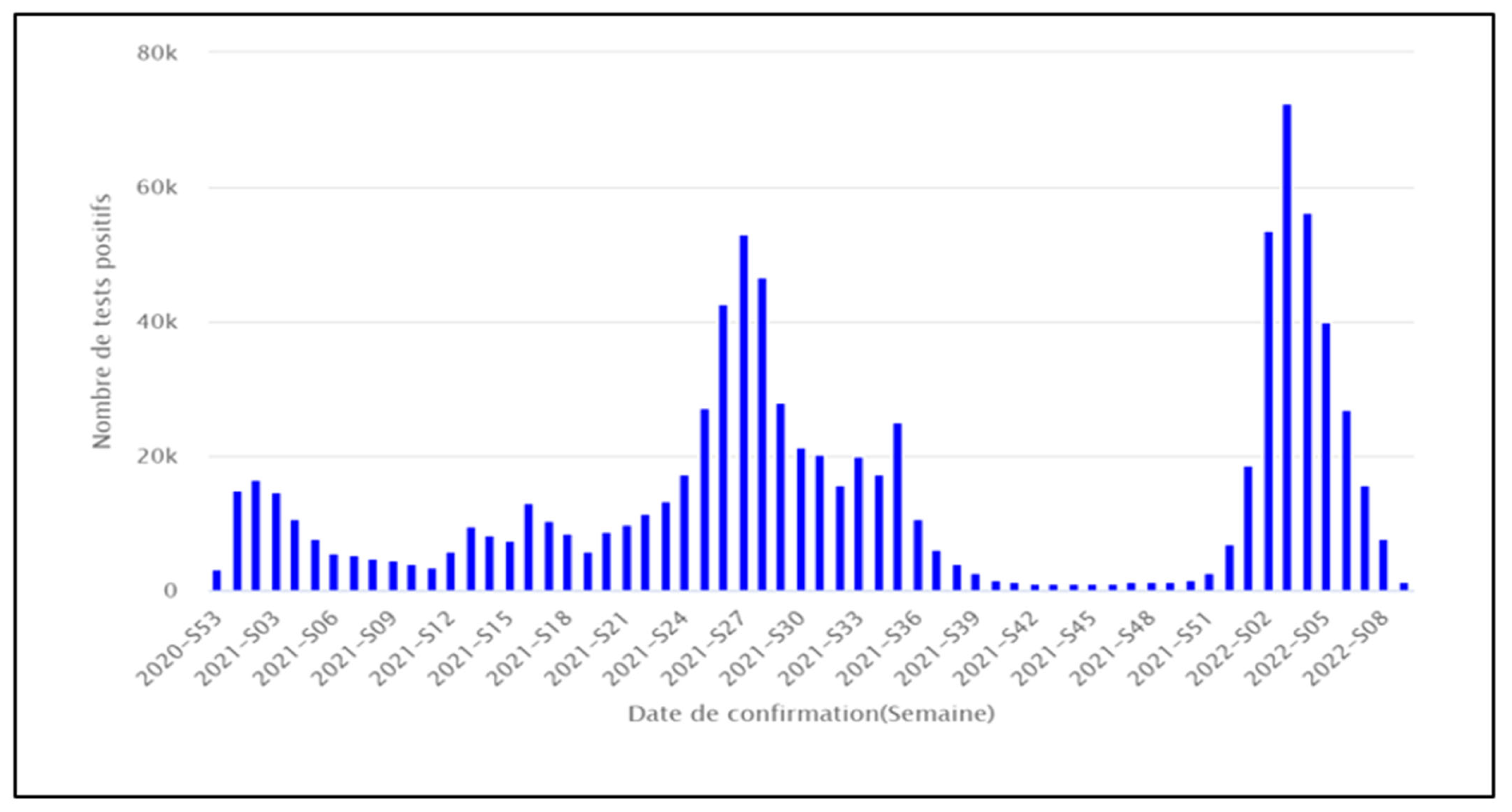
3.1. Surveillance Sequencing Data of SARS-CoV-2
3.2. Results of the Internal Audit of the Tunisian Strategy of SARS-CoV-2 Genomic Surveillance
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petersen, E.; Ntoumi, F.; Hui, D.S.; Abubakar, A.; Kramer, L.D.; Obiero, C.; Tambyah, P.A.; Blumberg, L.; Yapi, R.; Al-Abri, S. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B.1.1.529)—Highlights Africa’s research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int. J. Infect. Dis. 2022, 114, 268–272. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. COVID-19 Weekly Epidemiological Update. Available online: https://apps.who.int/iris/bitstream/handle/10665/336478/nCoV-weekly-sitrep01Nov20-eng.pdf (accessed on 16 August 2022).
- World Health Organization. Genomic Sequencing of SARS-CoV-2: A Guide to Implementation for Maximum Impact on Public Health; World Health Organization: Geneva, Switzerland, 2021; 80 p. [Google Scholar]
- Chen, Z.; Azman, A.S.; Chen, X.; Zou, J.; Tian, Y.; Sun, R.; Xu, X.; Wu, Y.; Lu, W.; Ge, S.; et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat. Genet. 2022, 54, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.; Spott, R.; Hölzer, M.; Kühnert, D.; Fuchs, S.; Lohde, M.; Marquet, M.; Viehweger, A.; Rimek, D.; Pletz, M.W. Molecular epidemiology of SARS-CoV-2—A regional to global perspective. medRxiv 2021. [Google Scholar] [CrossRef]
- Abid, S.; Ferjani, S.; El Moussi, A.; Ferjani, A.; Nasr, M.; Landolsi, I.; Saidi, K.; Gharbi, H.; Letaief, H.; Hechaichi, A.; et al. Assessment of sample pooling for SARS-CoV-2 molecular testing for screening of asymptomatic persons in Tunisia. Diagn. Microbiol. Infect. Dis. 2020, 98, 115125. [Google Scholar] [CrossRef] [PubMed]
- Eurosurveillance. Economic Evaluation of Whole Genome Sequencing for Pathogen Identification and Surveillance—Results of Case Studies in Europe and the Americas 2016 to 2019. Available online: https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.9.1900606?crawler=true (accessed on 10 August 2022).
- Ladhani, S.N.; Ireland, G.; Baawuah, F.; Beckmann, J.; Okike, I.O.; Ahmad, S.; Garstang, J.; Brent, A.J.; Brent, B.; Aiano, F.; et al. Emergence of SARS-CoV-2 Alpha (B.1.1.7) variant, infection rates, antibody seroconversion and seroprevalence rates in secondary school students and staff: Active prospective surveillance, December 2020 to March 2021, England. J. Infect. 2021, 83, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Brito, A.F.; Semenova, E.; Dudas, G.; Hassler, G.W.; Kalinich, C.; Kraemer, M.; Ho, J.; Tegally, H.; Githinji, G.; Agoti, C.; et al. Global disparities in SARS-CoV-2 genomic surveillance. medRxiv 2021. [Google Scholar] [CrossRef] [PubMed]

| Areas of Requirements | Number of Requirements | Requirements |
|---|---|---|
| Sampling | 3 | 1-Random samples, representative of the geographic and demographic distribution of SARS-CoV-2 infections; |
| 2-Targeted sampling, focusing on particular subsets of cases associated with public health risks; | ||
| 3-Outbreaks, alerts, or other unusual events. | ||
| Data collection/analysis/sharing and decision making | 6 | 1-All reported sequences should be accompanied by metadata; |
| 2-Rapid sharing of genomic sequence information in public databases; | ||
| 3-Comprehensive epidemiological and clinical data can be linked to sequence information; | ||
| 4-Matching vaccine data to variant types will help determine the ability of a variant to evade the immune system response; | ||
| 5-Epidemiological studies of variants should focus on settings where the potential for generalization is high and the results are likely to be broadly relevant; | ||
| 6-The main objectives of an investigation of the first cases and their close contacts are to obtain a description or estimate of such things as the clinical picture of SARS-CoV-2 infection, and the associated disease course; the secondary infection rate (SIR) and secondary clinical attack rate of SARS-CoV-2 infection in close contacts; the serial interval of SARS-CoV-2 infection; the proportion of symptomatic cases among COVID-19 cases; and the identification of possible transmission routes. | ||
| Reporting of surveillance data on SARS-CoV-2 variants | 1 | Rapid sharing of information on variants is an essential element in understanding and eradicating SARS-CoV-2 globally. |
| Partnership | 1 | Countries with limited sequencing capacity are strongly encouraged to facilitate access to regional and international sequencing partnerships or to increase their capacity through existing sequencing systems or laboratory networks. |
| Ethical considérations | 1 | All ethical considerations related to genomic sequencing, sequence sharing, and related metadata will be discussed by the different stakeholders, a dossier will be prepared and submitted to an ethics committee. |
| Areas of Requirements | Requirements | Level of Compliance | Problem |
|---|---|---|---|
| Sampling | 1-Random samples, representative of the geographic and demographic distribution of SARS-CoV-2 infections; | Not met | Random sample was selected every week and problems in sample conservation (bank of samples) or in sample transport were detected |
| 2-Targeted sampling, focusing on particular subsets of cases associated with public health risks; | Partially met | Applied but in a partial way | |
| 3-Outbreaks, alerts, or other unusual events. | Partially met | Not systematically applied | |
| Data collection/ Analysis/ sharing and decision making | 1-All reported footage should be accompanied by metadata; | Not met | Lack of information about patients personal data |
| 2-Rapid sharing of genomic sequence information in public databases; | Partially met | Not systematically applied | |
| 3-Comprehensive epidemiological and clinical data can be linked to sequence information; | Partially met | Patient’s Information was not complete | |
| 4-Matching vaccine data to variant types will help determine the ability of a variant to evade the immune system response; | Not met | We did not have any vaccination data base or information. | |
| 5-Epidemiological studies of variants should focus on settings where the potential for generalization is high, and the results are likely to be broadly relevant; | Not met | Problem in data disponibility to conduct studies | |
| 6-The main objectives of an investigation of the first cases and their close contacts are to obtain a description or estimate of such things as the clinical picture of SARS-CoV-2 infection, and the associated disease course; the secondary infection rate (SIR) and secondary clinical attack rate of SARS-CoV-2 infection in close contacts; the serial interval of SARS-CoV-2 infection; the proportion of symptomatic cases among COVID-19 cases; and the identification of possible transmission routes. | Not met | Lack of information | |
| Reporting of surveillance data on SARS-CoV-2 variants | 1. Rapid sharing of information on variants is an essential element in understanding and eradicating SARS-CoV-2 globally. | Not met | Lack of sharing and publications |
| Partnership | 1. Countries with limited sequencing capacity are strongly encouraged to facilitate access to regional and international sequencing partnerships or to increase their capacity through existing sequencing systems or laboratory networks. | Fully met | There were partners, but partnership projects faced many difficulties |
| Ethical considerations | 1. All ethical considerations related to genomic sequencing, sequence sharing, and related metadata will be discussed by the different stakeholders, a folder will be prepared and submitted to an ethics committee. | Not met | Not applied |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neffati, A.; Safer, M.; Kalai, W.; Hechaichi, A.; Dhaouadi, S.; Letaief, H.; Aichouch, C.; Bouabid, L.; Darouiche, S.; El Mili, N.; et al. Genomic Surveillance of SARS-CoV-2: Data Analysis and Assessment of Tunisian Strategy from January 2021 to February 2022. Epidemiologia 2024, 5, 80-89. https://doi.org/10.3390/epidemiologia5010005
Neffati A, Safer M, Kalai W, Hechaichi A, Dhaouadi S, Letaief H, Aichouch C, Bouabid L, Darouiche S, El Mili N, et al. Genomic Surveillance of SARS-CoV-2: Data Analysis and Assessment of Tunisian Strategy from January 2021 to February 2022. Epidemiologia. 2024; 5(1):80-89. https://doi.org/10.3390/epidemiologia5010005
Chicago/Turabian StyleNeffati, Arwa, Mouna Safer, Wissal Kalai, Aicha Hechaichi, Sonia Dhaouadi, Hajer Letaief, Chaima Aichouch, Leila Bouabid, Sondes Darouiche, Nawel El Mili, and et al. 2024. "Genomic Surveillance of SARS-CoV-2: Data Analysis and Assessment of Tunisian Strategy from January 2021 to February 2022" Epidemiologia 5, no. 1: 80-89. https://doi.org/10.3390/epidemiologia5010005
APA StyleNeffati, A., Safer, M., Kalai, W., Hechaichi, A., Dhaouadi, S., Letaief, H., Aichouch, C., Bouabid, L., Darouiche, S., El Mili, N., Triki, H., Boutiba, I., Mastouri, M., Berrajah, L. F., & Bouafif Ben Alaya, N. (2024). Genomic Surveillance of SARS-CoV-2: Data Analysis and Assessment of Tunisian Strategy from January 2021 to February 2022. Epidemiologia, 5(1), 80-89. https://doi.org/10.3390/epidemiologia5010005






