Rapid and Convenient Quantitative Analysis of SARS-CoV-2 RNA in Serous Saliva with a Direct PCR Method
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Collection and Pretreatment of Saliva
2.3. Real-Time PCR Analysis
2.4. Linear Regression Analyses of the N1 and N2 Analysis in RNase-Free Water and Non-Diluted (Serous) Saliva
2.5. Detection of SARS-CoV-2 RNA at Low Concentrations
2.6. Statistical Analysis
3. Results
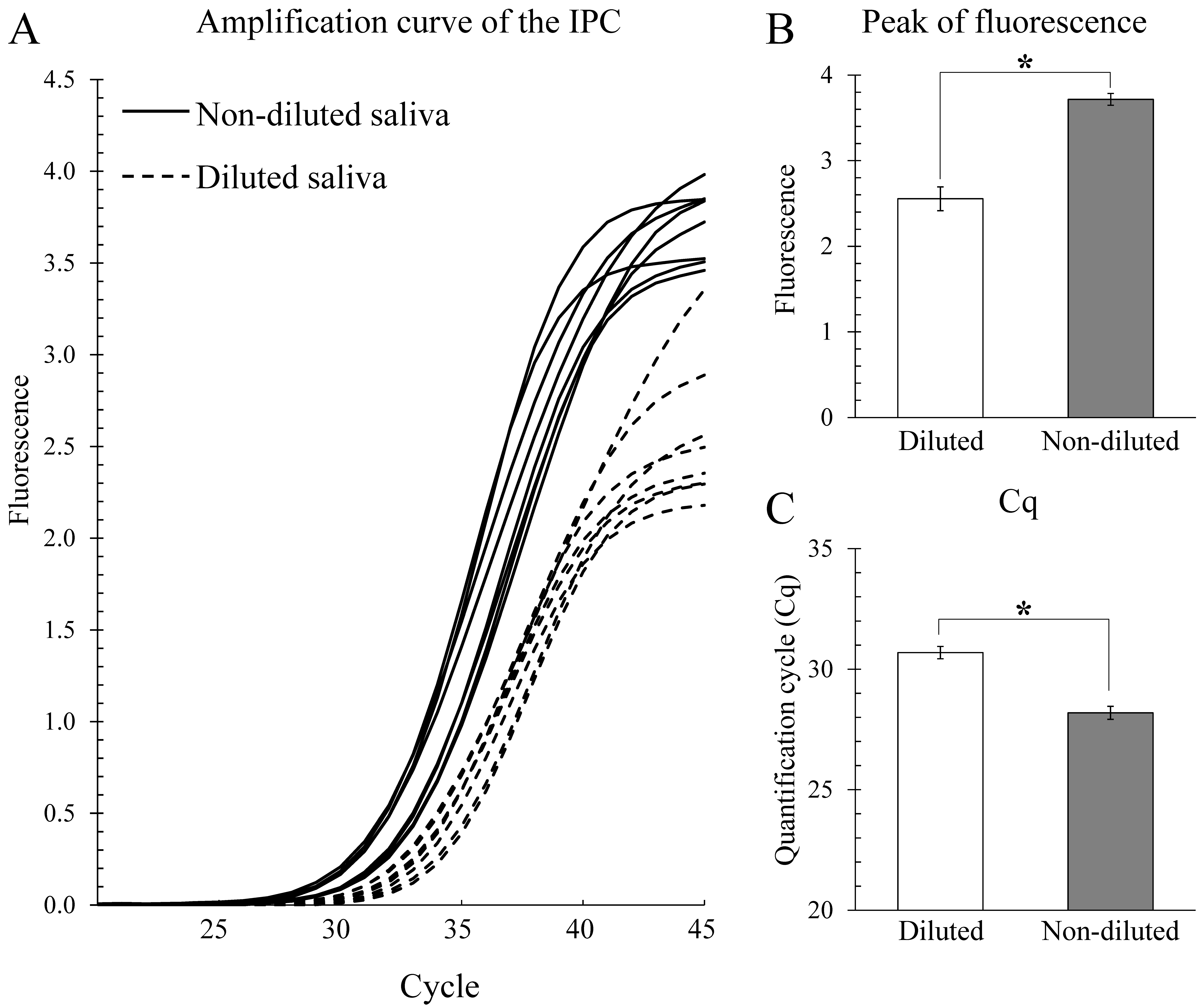
3.1. Amplification of IPC in Diluted and Non-Diluted Saliva Samples
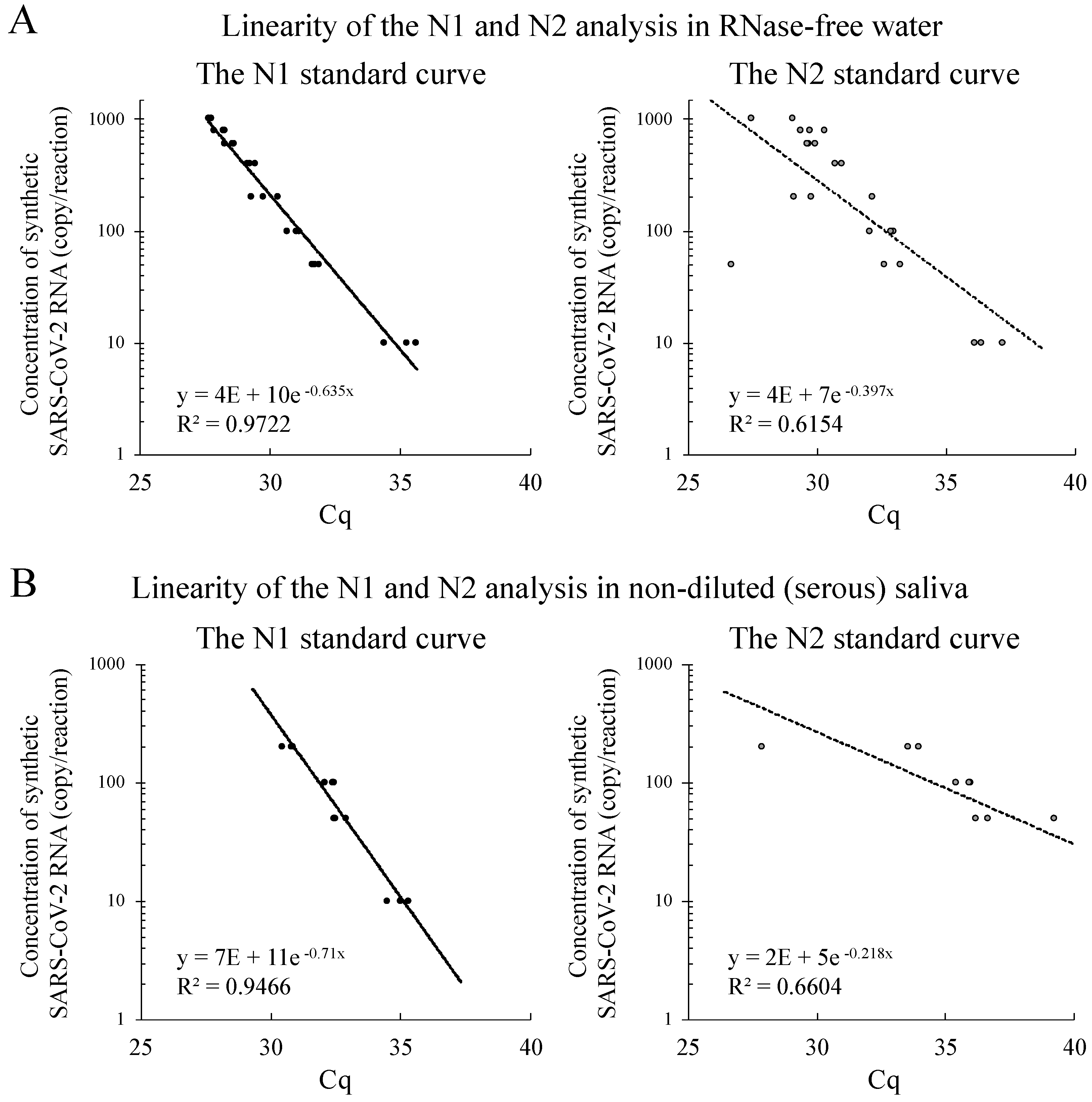
3.2. Linear Regression Analysis of N1 and N2 Amplifications
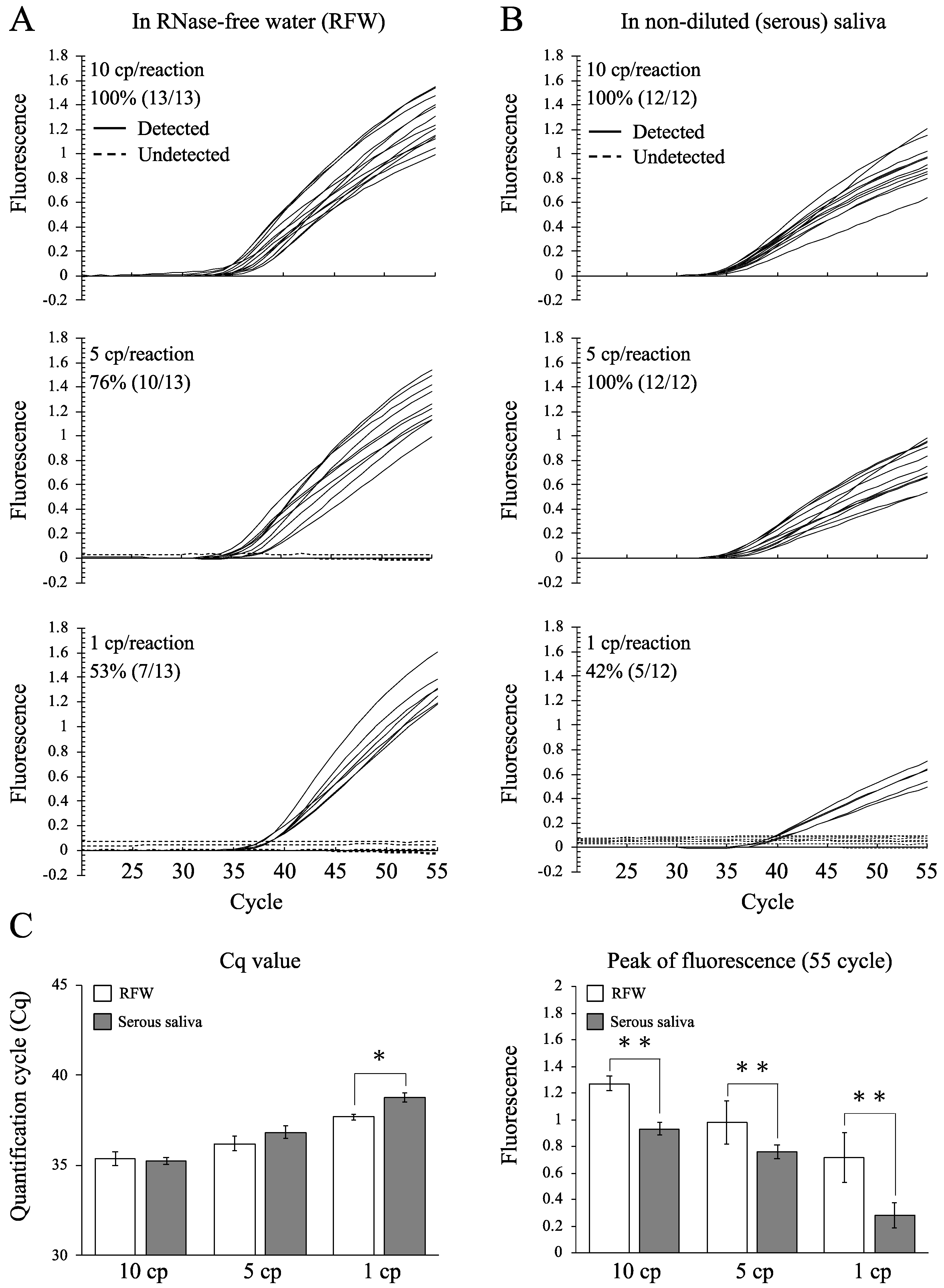
3.3. Detection Rates of N1 in Samples Containing Low Concentrations of SARS-CoV-2 RNA
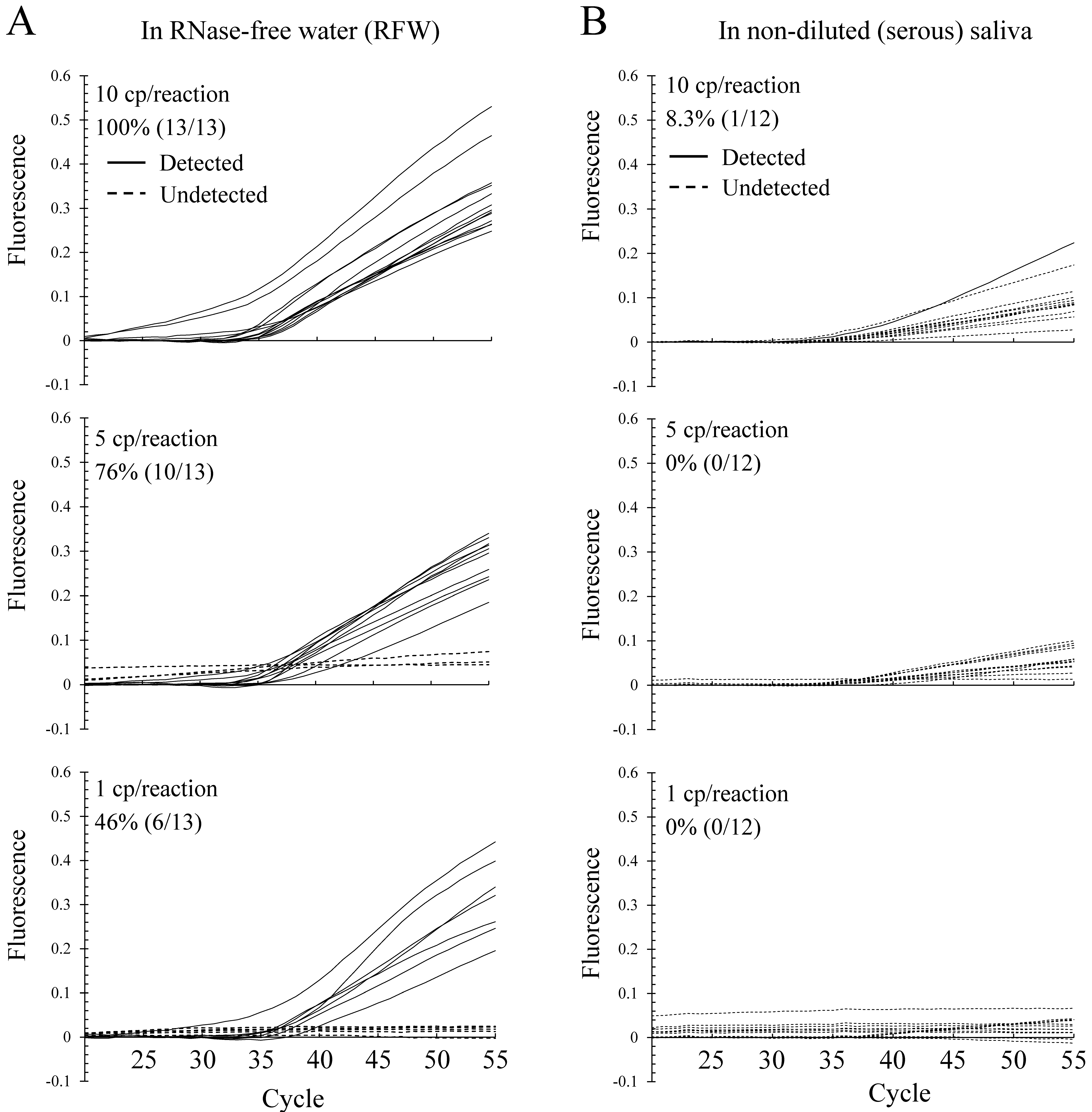
3.4. Detection Rates of N2 in Samples Containing Low Concentrations of SARS-CoV-2 RNA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gandhi, M.; Yokoe, D.S.; Havlir, D.V. Asymptomatic transmission, the Achilles’ heel of current strategies to control COVID-19. N. Engl. J. Med. 2020, 382, 2158–2160. [Google Scholar] [CrossRef] [PubMed]
- Oran, D.P.; Topol, E.J. Prevalence of asymptomatic SARS-CoV-2 infection: A narrative review. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef]
- Yamazaki, W.; Matsumura, Y.; Thongchankaew-Seo, U.; Yamazaki, Y.; Nagao, M. Development of a point-of-care test to detect SARS-CoV-2 from saliva which combines a simple RNA extraction method with colorimetric reverse transcription loop-mediated isothermal amplification detection. J. Clin. Virol. 2021, 136, 104760. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, Z.; Wang, X.; Wang, Y.; Wang, Y.; Wang, G.; Ren, L.; Li, J. Comparison of three TaqMan real-time reverse transcription-PCR assays in detecting SARS-CoV-2. J. Virol. Methods 2021, 288, 114030. [Google Scholar] [CrossRef]
- Azmi, I.; Faizan, M.I.; Kumar, R.; Raj Yadav, S.; Chaudhary, N.; Kumar Singh, D.; Butola, R.; Ganotra, A.; Joshi, G.D.; Jhingan, G.D.; et al. Saliva-based RNA extraction-free workflow integrated with Cas13a for SARS-CoV-2 detection. Front. Cell. Infect. Microbiol. 2021, 11, 632646. [Google Scholar] [CrossRef]
- Jiang, W.; Yu, H.T.; Zhao, K.; Zhang, Y.; Du, H.; Wang, P.Z.; Bai, X.F. Quantification of Hantaan virus with a SYBR green I-based one-step qRT-PCR assay. PLoS ONE. 2013, 8, e81525. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, W.; Zhu, Z.; Xia, X. Laboratory verification of an RT-PCR assay for SARS-CoV-2. J. Clin. Lab. Anal. 2020, 34, e23507. [Google Scholar] [CrossRef] [PubMed]
- Han, M.S.; Byun, J.H.; Cho, Y.; Rim, J.H. RT-PCR for SARS-CoV-2: Quantitative versus qualitative. Lancet Infect. Dis. 2021, 21, 165. [Google Scholar] [CrossRef]
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for di-agnostic COVID-19 tests. Nat. Rev. Microbiol. 2021, 19, 171–183. [Google Scholar] [CrossRef] [PubMed]
- McCormick-Baw, C.; Morgan, K.; Gaffney, D.; Cazares, Y.; Jaworski, K.; Byrd, A.; Molberg, K.; Cavuoti, D. Saliva as an alternate specimen source for detection of SARS-CoV-2 in sympto-matic patients using Cepheid Xpert Xpress SARS-CoV-2. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Fujisawa, S.; Nakakubo, S.; Kamada, K.; Yamashita, Y.; Fukumoto, T.; Sato, K.; Oguri, S.; Taki, K.; Senjo, H.; et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 2020, 81, e145–e147. [Google Scholar] [CrossRef]
- Yoon., J.G.; Yoon, J.; Song, J.Y.; Yoon, S.Y.; Lim, C.S.; Seong, H.; Noh, J.Y.; Cheong, H.J.; Kim, W.J. Clinical significance of a high SARS-CoV-2 viral load in the saliva. J. Korean Med. Sci. 2020, 35, e195. [Google Scholar] [CrossRef]
- The Gene Page for Angiotensin I Converting Enzyme 2 (ACE2: ENSG00000130234.10). The Gen-Otype-Tissue Expression (GTEx) Portal Website. Available online: https://www.gtexportal.org/home/gene/ACE2#geneExpression (accessed on 21 June 2021).
- Xu, J.; Li, Y.; Gan, F.; Du, Y.; Yao, Y. Salivary Glands: Potential Reservoirs for COVID-19 Asymp-tomatic Infection. J. Dent. Res. 2020, 99, 989. [Google Scholar] [CrossRef] [Green Version]
- Fukumoto, T.; Iwasaki, S.; Fujisawa, S.; Hayasaka, K.; Sato, K.; Oguri, S.; Taki, K.; Nakakubo, S.; Kamada, K.; Yamashita, Y.; et al. Efficacy of a novel SARS-CoV-2 detection kit without RNA extraction and purification. Int. J. Infect. Dis. 2020, 98, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, re-verse transcription-loop-mediated isothermal amplification, and a rapid antigen test to di-agnose COVID-19. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Research Use only 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primers and Probes In Viral Testing with the CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 21 June 2021).
- Rangan, R.; Zheludev, I.N.; Das, R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Lawler, D.M. Turbidity, Turbidimetry, and Nephelometry. In Encyclopedia of Analytical Science, 3rd ed.; Worsfold, P., Poole, C., Townshend, A., Miró, M., Eds.; Academic Press: Oxford, UK, 2016; pp. 152–163. [Google Scholar] [CrossRef]
| Steps | Reactions | Cycles |
|---|---|---|
| Reverse transcription | 42 °C for 600 s | 1 |
| Preincubation | 95 °C for 60 s | 1 |
| 2-Step amplification | 95 °C for 5 s | 45 or 55 |
| 60 °C for 30 s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deura, C.; Nakayama, K. Rapid and Convenient Quantitative Analysis of SARS-CoV-2 RNA in Serous Saliva with a Direct PCR Method. Epidemiologia 2021, 2, 305-314. https://doi.org/10.3390/epidemiologia2030023
Deura C, Nakayama K. Rapid and Convenient Quantitative Analysis of SARS-CoV-2 RNA in Serous Saliva with a Direct PCR Method. Epidemiologia. 2021; 2(3):305-314. https://doi.org/10.3390/epidemiologia2030023
Chicago/Turabian StyleDeura, Chikaya, and Kenji Nakayama. 2021. "Rapid and Convenient Quantitative Analysis of SARS-CoV-2 RNA in Serous Saliva with a Direct PCR Method" Epidemiologia 2, no. 3: 305-314. https://doi.org/10.3390/epidemiologia2030023
APA StyleDeura, C., & Nakayama, K. (2021). Rapid and Convenient Quantitative Analysis of SARS-CoV-2 RNA in Serous Saliva with a Direct PCR Method. Epidemiologia, 2(3), 305-314. https://doi.org/10.3390/epidemiologia2030023





