Interactions between 14-3-3 Proteins and Actin Cytoskeleton and Its Regulation by microRNAs and Long Non-Coding RNAs in Cancer
Abstract
1. Introduction
3. Oncogenic Role of 14-3-3α/β in Cancer
4. Oncogenic Role of 14-3-3 Eta
5. Opposing Role of 14-3-3ε in Cancer
6. Oncogenic Role of 14-3-3 Gamma
7. Opposing Roles of 14-3-3 Sigma in Cancer
8. Oncogeinc Role of 14-3-3 Theta in Cancer
9. Oncogenic Roles of 14-3-3 Zeta/Delta in Cancer
10. Interactions between 14-3-3 Proteins and Actin Cytoskeleton in Cancer
11. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pennington, K.L.; Chan, T.Y.; Torres, M.P.; Andersen, J.L. The dynamic and stress-adaptive signaling hub of 14-3-3: Emerging mechanisms of regulation and context-dependent protein–protein interactions. Oncogene 2018, 37, 5587–5604. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Daut, J.; Schwappach, B. Membrane Proteins as 14-3-3 Clients in Functional Regulation and Intracellular Transport. Physiology 2011, 26, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Chaudhri, M.; Scarabel, M.; Aitken, A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem. Biophys. Res. Commun. 2002, 300, 679–685. [Google Scholar] [CrossRef]
- Fan, X.; Cui, L.; Zeng, Y.; Song, W.; Gaur, U.; Yang, M. 14-3-3 Proteins Are on the Crossroads of Cancer, Aging, and Age-Related Neurodegenerative Disease. Int. J. Mol. Sci. 2019, 20, 3518. [Google Scholar] [CrossRef]
- Sluchanko, N.; Gusev, N.B. Oligomeric structure of 14-3-3 protein: What do we know about monomers? FEBS Lett. 2012, 586, 4249–4256. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.B. How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2001, 513, 53–57. [Google Scholar] [CrossRef]
- Johnson, L.N.; Barford, D. The Effects of Phosphorylation on the Structure and Function of Proteins. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 199–232. [Google Scholar] [CrossRef]
- Pair, F.S.; Yacoubian, T.A. 14-3-3 Proteins: Novel Pharmacological Targets in Neurodegenerative Diseases. Trends Pharmacol. Sci. 2021, 42, 226–238. [Google Scholar] [CrossRef]
- Cho, E. Emerging roles of 14-3-3γ in the brain disorder. BMB Rep. 2020, 53, 500–511. [Google Scholar] [CrossRef]
- Silhan, J.; Obšilová, V.; Vecer, J.; Herman, P.; Sulc, M.; Teisinger, J.; Obsil, T. 14-3-3 Protein C-terminal Stretch Occupies Ligand Binding Groove and Is Displaced by Phosphopeptide Binding. J. Biol. Chem. 2004, 279, 49113–49119. [Google Scholar] [CrossRef]
- Obsilova, V.; Herman, P.; Vecer, J.; Sulc, M.; Teisinger, J.; Obsil, T. 14-3-3zeta C-terminal Stretch Changes Its Conformation upon Ligand Binding and Phosphorylation at Thr232. J. Biol. Chem. 2004, 279, 4531–4540. [Google Scholar] [CrossRef] [PubMed]
- Mackintosh, C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes Carol Mackintosh. Biochem. J. 2004, 381, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Bagwan, N.; El Ali, H.H.; Lundby, A. Proteome-wide profiling and mapping of post translational modifications in human hearts. Sci. Rep. 2021, 11, 2184. [Google Scholar] [CrossRef]
- Obsilova, V.; Obsil, T. The 14-3-3 Proteins as Important Allosteric Regulators of Protein Kinases. Int. J. Mol. Sci. 2020, 21, 8824. [Google Scholar] [CrossRef]
- Liu, T.-A.; Jan, Y.-J.; Ko, B.-S.; Chen, S.-C.; Liang, S.-M.; Hung, Y.-L.; Hsu, C.; Shen, T.-L.; Lee, Y.-M.; Chen, P.-F.; et al. Increased Expression of 14-3-3β Promotes Tumor Progression and Predicts Extrahepatic Metastasis and Worse Survival in Hepatocellular Carcinoma. Am. J. Pathol. 2011, 179, 2698–2708. [Google Scholar] [CrossRef]
- Lin, H.; Jiao, X.; Yu, B.; Du, J.; Xu, H.; Dong, A.; Wan, C. Clinical significance of serum 14-3-3 beta in patients with hepatocellular carcinoma. Cancer Biomark. 2017, 20, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Matsumoto, K.; Minamida, S.; Hirayama, T.; Fujita, T.; Kodera, Y.; Iwamura, M. Incremental Expression of 14-3-3 Protein Beta/Alpha in Urine Correlates with Advanced Stage and Poor Survival in Patients with Clear Cell Renal Cell Carcinoma. Asian Pac. J. Cancer Prev. 2016, 17, 1399–1404. [Google Scholar] [CrossRef]
- Minamida, S.; Iwamura, M.; Kodera, Y.; Kawashima, Y.; Tabata, K.; Matsumoto, K.; Fujita, T.; Satoh, T.; Maeda, T.; Baba, S. 14-3-3 Protein beta/alpha as a urinary biomarker for renal cell carcinoma: Proteomic analysis of cyst fluid. Anal. Bioanal. Chem. 2011, 401, 245–252. [Google Scholar] [CrossRef]
- Hu, X.; Bao, M.; Huang, J.; Zhou, L.; Zheng, S. Identification and Validation of Novel Biomarkers for Diagnosis and Prognosis of Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 541479. [Google Scholar] [CrossRef]
- Sugiyama, A.; Miyagi, Y.; Komiya, Y.; Kurabe, N.; Kitanaka, C.; Kato, N.; Nagashima, Y.; Kuchino, Y.; Tashiro, F. Forced expression of antisense 14-3-3 RNA suppresses tumor cell growth in vitro and in vivo. Carcinogenesis 2003, 24, 1549–1559. [Google Scholar] [CrossRef]
- Takihara, Y.; Matsuda, Y.; Hara, J. Role of the beta isoform of 14-3-3 proteins in cellular proliferation and oncogenic transformation. Carcinogenesis 2000, 21, 2073–2077. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Jan, Y.-J.; Ko, B.-S.; Wu, Y.-J.; Wu, Y.-J.; Liou, J.-Y. Prognostic Significance of 14-3-3ε, Aldo-keto Reductase Family 1 B10 and Metallothionein-1 in Hepatocellular Carcinoma. Anticancer Res. 2018, 38, 6855–6863. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Akiyama, H.; Sakumoto, R.; Tashiro, F. FBI1/Akirin2 promotes tumorigenicity and metastasis of Lewis lung carcinoma cells. Biochem. Biophys. Res. Commun. 2014, 444, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Okayama, A.; Miyagi, Y.; Oshita, F.; Nishi, M.; Nakamura, Y.; Nagashima, Y.; Akimoto, K.; Ryo, A.; Hirano, H. Proteomic Analysis of Proteins Related to Prognosis of Lung Adenocarcinoma. J. Proteome Res. 2014, 13, 4686–4694. [Google Scholar] [CrossRef]
- Xu, C.; Du, Z.; Ren, S.; Liang, X.; Li, H. MiR-129-5p sensitization of lung cancer cells to etoposide-induced apoptosis by reducing YWHAB. J. Cancer 2020, 11, 858–866. [Google Scholar] [CrossRef]
- Liu, L.; Liu, Z.; Wang, H.; Chen, L.; Ruan, F.; Zhang, J.; Hu, Y.; Luo, H.; Wen, S. 14-3-3β exerts glioma-promoting effects and is associated with malignant progression and poor prognosis in patients with glioma. Exp. Ther. Med. 2017, 15, 2381–2387. [Google Scholar] [CrossRef]
- Cao, L.; Lei, H.; Chang, M.-Z.; Liu, Z.-Q.; Bie, X.-H. Down-regulation of 14-3-3β exerts anti-cancer effects through inducing ER stress in human glioma U87 cells: Involvement of CHOP–Wnt pathway. Biochem. Biophys. Res. Commun. 2015, 462, 389–395. [Google Scholar] [CrossRef]
- Kang, C.-M.; Bai, H.-L.; Li, X.-H.; Huang, R.-Y.; Zhao, J.-J.; Dai, X.-Y.; Zheng, L.; Qiu, Y.-R.; Hu, Y.-W.; Wang, Q. The binding of lncRNA RP11-732M18.3 with 14-3-3 β/α accelerates p21 degradation and promotes glioma growth. eBioMedicine 2019, 45, 58–69. [Google Scholar] [CrossRef]
- Yang, Y.-F.; Lee, Y.; Wang, Y.; Wang, C.; Hou, M.; Yuan, S.F. YWHAE promotes proliferation, metastasis, and chemoresistance in breast cancer cells. Kaohsiung J. Med. Sci. 2019, 35, 408–416. [Google Scholar] [CrossRef]
- Jia, S.; Chen, G.; Liang, Y.; Liang, X.; Meng, C.Y. GCH1-regulated miRNAs are potential targets for microglial activation in neuropathic pain. Biosci. Rep. 2021, 41, BSR20210051. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Q.; Zhang, K.; Wang, F.; Qiao, X.; Cui, J. Circ SMARCA5 Inhibited Tumor Metastasis by Interacting with SND1 and Downregulating the YWHAB Gene in Cervical Cancer. Cell Transplant. 2021, 30, 963689720983786. [Google Scholar] [CrossRef]
- Wang, Z.; Nesland, J.M.; Suo, Z.; Trope, C.G.; Holm, R. The Prognostic Value of 14-3-3 Isoforms in Vulvar Squamous Cell Carcinoma Cases: 14-3-3β and ε Are Independent Prognostic Factors for These Tumors. PLoS ONE 2011, 6, e24843. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.N.; Sharma, N. Quantitative SWATH-Based Proteomic Profiling for Identification of Mechanism-Driven Diagnostic Biomarkers Conferring in the Progression of Metastatic Prostate Cancer. Front. Oncol. 2020, 10, 00493. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, P.; Mondal, A.K.; Bloomer, C.; Fulzele, S.; Jones, K.; Ananth, S.; Gahlay, G.K.; Heneidi, S.; Rojiani, A.M.; Kota, V.; et al. Identification and Clinical Validation of a Novel 4 Gene-Signature with Prognostic Utility in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 3818. [Google Scholar] [CrossRef]
- Wang, T.; Huang, X.-Y.; Zheng, S.-J.; Liu, Y.-Y.; Chen, S.-S.; Ren, F.; Lu, J.; Duan, Z.-P.; Liu, M. Serum Anti-14-3-3 Zeta Autoantibody as a Biomarker for Predicting Hepatocarcinogenesis. Front. Oncol. 2021, 11, 733680. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Jiang, Y.; Ji, X.; Guan, X.; Lin, Q.; Ma, L. Identification of novel prognostic genes of triple-negative breast cancer using meta-analysis and weighted gene co-expressed network analysis. Ann. Transl. Med. 2021, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.-J.; Qian, G.-Y.; Yang, J.-M.; Lee, J.; Park, Y.-D. Detection of Melanogenesis and Anti-Apoptosis-Associated Melanoma Factors: Array CGH and PPI Mapping Integrating Study. Protein Pept. Lett. 2021, 28, 1408–1424. [Google Scholar] [CrossRef]
- Barbosa-Silva, A.; Magalhães, M.; Da Silva, G.F.; Da Silva, F.A.B.; Carneiro, F.R.G.; Carels, N. A Data Science Approach for the Identification of Molecular Signatures of Aggressive Cancers. Cancers 2022, 14, 2325. [Google Scholar] [CrossRef]
- Wang, Y.; Jacobs, C.; E Hook, K.; Duan, H.; Booher, R.N.; Sun, Y. Binding of 14-3-3beta to the carboxyl terminus of Wee1 increases Wee1 stability, kinase activity, and G2-M cell population. Cell Growth Differ. 2000, 11, 211–219. [Google Scholar]
- Han, D.C.; Rodriguez, L.G.; Guan, J.-L. Identification of a novel interaction between integrin β1 and 14-3-3β. Oncogene 2001, 20, 346–357. [Google Scholar] [CrossRef]
- Jasinski-Bergner, S.; Stehle, F.; Gonschorek, E.; Kalich, J.; Schulz, K.; Huettelmaier, S.; Braun, J.; Seliger, B. Identification of 14-3-3β Gene as a Novel miR-152 Target Using a Proteome-based Approach. J. Biol. Chem. 2014, 289, 31121–31135. [Google Scholar] [CrossRef] [PubMed]
- Perdigão-Henriques, R.; Petrocca, F.; Altschuler, G.; Thomas, M.P.; LE, T.N.M.; Tan, S.M.; Hide, W.; A Lieberman, J. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 2015, 35, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Wang, H.; Wang, H.; Wu, X.; Yang, L.; Wang, C.; Li, X.; Jin, Y.; Li, M.; Wang, L.; et al. Circular RNA Circ_0006282 Promotes Cell Proliferation and Metastasis in Gastric Cancer by Regulating MicroRNA-144-5p/Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein β Axis. Cancer Manag. Res. 2021, 13, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Park, G.-Y.; Han, J.Y.; Han, Y.K.; Kim, S.D.; Kim, J.S.; Jo, W.S.; Chun, S.H.; Jeong, D.H.; Lee, C.-W.; Yang, K.; et al. 14-3-3 eta depletion sensitizes glioblastoma cells to irradiation due to enhanced mitotic cell death. Cancer Gene Ther. 2014, 21, 158–163. [Google Scholar] [CrossRef]
- Lee, C.G.; Park, G.-Y.; Han, Y.K.; Lee, J.H.; Chun, S.H.; Park, H.-Y.; Lim, K.-H.; Kim, E.-G.; Choi, Y.-J.; Yang, K. Roles of 14-3-3η in mitotic progression and its potential use as a therapeutic target for cancers. Oncogene 2012, 32, 1560–1569. [Google Scholar] [CrossRef]
- Liang, S.; Singh, M.; Gam, L.-H. The Differential Expression of Aqueous Soluble Proteins in Breast Normal and Cancerous Tissues in Relation to Stage and Grade of Patients. J. Biomed. Biotechnol. 2010, 2010, 516469. [Google Scholar] [CrossRef][Green Version]
- Shen, J.; Jiang, F.; Yang, Y.; Huang, G.; Pu, F.; Liu, Q.; Chen, L.; Ju, L.; Lu, M.; Zhou, F.; et al. 14-3-3η is a novel growth-promoting and angiogenic factor in hepatocellular carcinoma. J. Hepatol. 2016, 65, 953–962. [Google Scholar] [CrossRef]
- Qiu, Y.; Shan, W.; Yang, Y.; Jin, M.; Dai, Y.; Yang, H.; Jiao, R.; Xia, Y.; Liu, Q.; Ju, L.; et al. Reversal of sorafenib resistance in hepatocellular carcinoma: Epigenetically regulated disruption of 14-3-3η/hypoxia-inducible factor-1α. Cell Death Discov. 2019, 5, 120. [Google Scholar] [CrossRef]
- Lin, J.-P.; Fan, Y.-K.; Liu, H.M. The 14-3-3η chaperone protein promotes antiviral innate immunity via facilitating MDA5 oligomerization and intracellular redistribution. PLoS Pathog. 2019, 15, e1007582. [Google Scholar] [CrossRef]
- Wu, Q.; Fan, H.; Lang, R.; Li, X.; Zhang, X.; Lv, S.; He, Q. Overexpression of 14-3-3σ Modulates Cholangiocarcinoma Cell Survival by PI3K/Akt Signaling. BioMed Res. Int. 2020, 2020, 3740418. [Google Scholar] [CrossRef]
- Davidson, B.; Holth, A.; Wang, Z.; Hellsylt, E.; Tropé, C.G.; Falkenthal, T.E.H.; Holm, R. Expression of 14-3-3 sigma and eta proteins is unrelated to survival in metastatic high-grade serous carcinoma. APMIS 2018, 126, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Titus, M.A.; Tan, J.-A.; Gregory, C.W.; Ford, O.H.; Subramanian, R.R.; Fu, H.; Wilson, E.M.; Mohler, J.L.; French, F.S. 14-3-3η Amplifies Androgen Receptor Actions in Prostate Cancer. Clin. Cancer Res. 2009, 15, 7571–7581. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-H.; Kikuchi, K.; Zheng, Y.; Noguchi, A.; Takahashi, H.; Nishida, T.; Masuda, S.; Yang, X.-H.; Takano, Y. Downregulation and translocation of nuclear ING4 is correlated with tumorigenesis and progression of head and neck squamous cell carcinoma. Oral Oncol. 2011, 47, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Q.; Zhang, M.; Luo, Y.; Fu, Y. Downregulation of nuclear ING3 expression and translocalization to cytoplasm promotes tumorigenesis and progression in head and neck squamous cell carcinoma (HNSCC). Histol. Histopathol. 2019, 35, 681–690. [Google Scholar] [CrossRef]
- Castañeda, A.; Serrano, C.; Hernández-Trejo, J.A.; Gutiérrez-Martínez, I.Z.; Montejo-López, W.; Gómez-Suárez, M.; Hernández-Ruiz, M.; Betanzos, A.; Candelario-Martínez, A.; Romo-Parra, H.; et al. pVHL suppresses Akt/β-catenin-mediated cell proliferation by inhibiting 14-3-3ζ expression. Biochem. J. 2017, 474, 2679–2689. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, H.; Shin, S.M.; Kim, H.K.; Ha, S.-A.; Cho, G.W.; Hur, S.Y.; Kim, T.E.; Kim, J.W. The bone morphogenetic protein antagonist gremlin 1 is overexpressed in human cancers and interacts with YWHAH protein. BMC Cancer 2006, 6, 74. [Google Scholar] [CrossRef]
- Wanzel, M.; Kleine-Kohlbrecher, D.; Herold, S.; Hock, A.; Berns, K.; Park, J.; Hemmings, B.; Eilers, M. Akt and 14-3-3η regulate Miz1 to control cell-cycle arrest after DNA damage. Nat. Cell Biol. 2004, 7, 30–41. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, Q.; Li, C.; Zhou, Y.; Zhao, S.; Hong, L.; Song, Q.; Yu, S.; Hu, C.; Wang, H.; et al. Enhancement of mitochondrial biogenesis and paradoxical inhibition of lactate dehydrogenase mediated by 14-3-3η in oncocytomas. J. Pathol. 2018, 245, 361–372. [Google Scholar] [CrossRef]
- Zhao, S.; Li, B.; Li, C.; Gao, H.; Miao, Y.; He, Y.; Wang, H.; Gong, L.; Li, D.; Zhang, Y.; et al. The Apoptosis Regulator 14-3-3η and Its Potential as a Therapeutic Target in Pituitary Oncocytoma. Front. Endocrinol. 2019, 10, 797. [Google Scholar] [CrossRef]
- Zhang, Z.; He, H.; Qiao, Y.; Huang, J.; Wu, Z.; Xu, P.; Yin, D.; He, M. Tanshinone IIA Pretreatment Protects H9c2 Cells against Anoxia/Reoxygenation Injury: Involvement of the Translocation of Bcl-2 to Mitochondria Mediated by 14-3-3η. Oxid. Med. Cell. Longev. 2018, 2018, 3583921. [Google Scholar] [CrossRef]
- Haonon, O.; Rucksaken, R.; Pinlaor, P.; Pairojkul, C.; Chamgramol, Y.; Intuyod, K.; Onsurathum, S.; Khuntikeo, N.; Pinlaor, S. Upregulation of 14-3-3 eta in chronic liver fluke infection is a potential diagnostic marker of cholangiocarcinoma. Proteom.-Clin. Appl. 2015, 10, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Zhen, J.; Jiao, K.; Yang, K.; Wu, M.; Zhou, Q.; Yang, B.; Xiao, W.; Hu, C.; Zhou, M.; Li, Z. The 14-3-3η/GSK-3β/β-catenin complex regulates EndMT induced by 27-hydroxycholesterol in HUVECs and promotes the migration of breast cancer cells. Cell Biol. Toxicol. 2020, 37, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, J.; Kang, Y.-L.; Woo, J.; Kim, Y.; Huh, J.; Park, J.-W. Ketohexokinase-A acts as a nuclear protein kinase that mediates fructose-induced metastasis in breast cancer. Nat. Commun. 2020, 11, 5436. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Yuan, D.; Sun, R.-J.; Zhu, J.-J.; Shan, N.-N. Significant reductions in apoptosis-related proteins (HSPA6, HSPA8, ITGB3, YWHAH, and PRDX6) are involved in immune thrombocytopenia. J. Thromb. Thrombolysis 2020, 51, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Han, Y.; Hu, S.; Cai, Y.; Yang, J.; Ren, S.; Zhao, Y.; Lu, T.; Zhou, X.; Wang, X. Circulating Exosomal MiR-107 Restrains Tumorigenesis in Diffuse Large B-Cell Lymphoma by Targeting 14-3-3η. Front. Cell Dev. Biol. 2021, 9, 667800. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, S.; Luo, Y.; Zhang, M.; Jiang, X.; Xiong, Y. IncRNA MAPKAPK5-AS1 promotes proliferation and migration of thyroid cancer cell lines by targeting miR-519e-5p/YWHAH. Eur. J. Histochem. 2020, 64, 3177. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, H.; Duan, Z.; Chen, X.; Ao, Z.; Chen, Y.; Ruan, Y.; Ni, M. miR-31-5p Regulates 14-3-3 ɛ to Inhibit Prostate Cancer 22RV1 Cell Survival and Proliferation via PI3K/AKT/Bcl-2 Signaling Pathway. Cancer Manag. Res. 2020, 12, 6679–6694. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, D.; Guo, P. Circ_0000144 acts as a miR-1178-3p decoy to promote cell malignancy and angiogenesis by increasing YWHAH expression in papillary thyroid cancer. J. Otolaryngol.-Head Neck Surg. 2022, 51, 28. [Google Scholar] [CrossRef]
- Leal, M.F.; Ribeiro, H.F.; Rey, J.A.; Pinto, G.R.; Smith, M.C.; Moreira-Nunes, C.A.; Assumpção, P.P.; Lamarão, L.M.; Calcagno, D.Q.; Montenegro, R.C.; et al. YWHAE silencing induces cell proliferation, invasion and migration through the up-regulation of CDC25B and MYC in gastric cancer cells: New insights about YWHAE role in the tumor development and metastasis process. Oncotarget 2016, 7, 85393–85410. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, F.; Ren, X.; Yang, Y.; Luo, J.; Yuan, J.; Yuan, J.; Tong, Q. RNA-Binding Protein COL14A1, TNS1, NUSAP1 and YWHAE Are Valid Biomarkers to Predict Peritoneal Metastasis in Gastric Cancer. Front. Oncol. 2022, 12, 830688. [Google Scholar] [CrossRef]
- Che, X.-H.; Chen, H.; Xu, Z.-M.; Shang, C.; Sun, K.-L.; Fu, W.-N. 14-3-3epsiloncontributes to tumour suppression in laryngeal carcinoma by affecting apoptosis and invasion. BMC Cancer 2010, 10, 306. [Google Scholar] [CrossRef] [PubMed]
- Cimino, D.; Fuso, L.; Sfiligoi, C.; Biglia, N.; Ponzone, R.; Maggiorotto, F.; Russo, G.; Cicatiello, L.; Weisz, A.; Taverna, D.; et al. Identification of new genes associated with breast cancer progression by gene expression analysis of predefined sets of neoplastic tissues. Int. J. Cancer 2008, 123, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, C.; Wang, S.; Hu, Y.; Jin, S.; Liu, O.; Gou, R.; Nie, X.; Liu, J.; Lin, B. YWHAE as an HE4 interacting protein can influence the malignant behaviour of ovarian cancer by regulating the PI3K/AKT and MAPK pathways. Cancer Cell Int. 2021, 21, 302. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Arbindi, H.; Zola, F.; Mattox, S.N.; Velasquez Zarate, L.; Munroe, J.; Heneidi, S.; Sharma, S.; Ghamande, S.; Kleven, D.T. Pleomorphic high grade endometrial stromal sarcoma with YWHAE gene amplification may be a novel variant with poor prognosis. Int. J. Clin. Exp. Pathol. 2022, 15, 72–78. [Google Scholar]
- Yang, Y.; Sang, Z.-Y.; Ma, J.; Zhu, Y.-P.; Wu, S.-F. KRAS, YWHAE, SP1 and MSRA as biomarkers in endometrial cancer. Transl. Cancer Res. 2021, 10, 1295–1312. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, W.; Zhao, Z.; Zhu, Y. JAZF1, YWHAE and BCOR gene translocation in primary extrauterine low-grade and high-grade endometrial stromal sarcomas. Histopathology 2021, 80, 809–819. [Google Scholar] [CrossRef]
- Mohammad, N.; Stewart, C.J.; Chiang, S.; Turashvili, G.; Dickson, B.C.; Ng, T.L.; Köbel, M.; McCluggage, W.G.; Croce, S.; Lee, C. p53 immunohistochemical analysis of fusion-positive uterine sarcomas. Histopathology 2020, 78, 805–813. [Google Scholar] [CrossRef]
- Ou, W.-B.; Lundberg, M.Z.; Zhu, S.; Bahri, N.; Kyriazoglou, A.; Xu, L.; Chen, T.; Mariño-Enriquez, A.; Fletcher, J.A. YWHAE-NUTM2 oncoprotein regulates proliferation and cyclin D1 via RAF/MAPK and Hippo pathways. Oncogenesis 2021, 10, 37. [Google Scholar] [CrossRef]
- Li, C.; Wang, C. LG-ESSs and HG-ESSs: Underlying molecular alterations and potential therapeutic strategies. J. Zhejiang Univ. Sci. B 2021, 22, 633–646. [Google Scholar] [CrossRef]
- Lee, C.-H.; Mariño-Enriquez, A.; Ou, W.; Zhu, M.; Ali, R.H.; Chiang, S.; Amant, F.; Gilks, C.B.; van de Rijn, M.; Oliva, E.; et al. The Clinicopathologic Features of YWHAE-FAM22 Endometrial Stromal Sarcomas: A histologically high-grade and clinically aggressive tumor. Am. J. Surg. Pathol. 2012, 36, 641–653. [Google Scholar] [CrossRef]
- Croce, S.; Hostein, I.; Ribeiro, A.; Garbay, D.; Velasco, V.; Stoeckle, E.; Guyon, F.; Floquet, A.; Neuville, A.; Coindre, J.-M.; et al. YWHAE rearrangement identified by FISH and RT-PCR in endometrial stromal sarcomas: Genetic and pathological correlations. Mod. Pathol. 2013, 26, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Ko, B.-S.; Jan, Y.-J.; Chang, T.-C.; Liang, S.-M.; Chen, S.-C.; Liu, T.-A.; Wu, Y.-M.; Wang, J.; Liou, J.-Y. Upregulation of Focal Adhesion Kinase by 14-3-3ε via NFκB Activation in Hepatocellular Carcinoma. Anti-Cancer Agents Med. Chem. 2013, 13, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Ko, B.-S.; Lai, I.-R.; Chang, T.-C.; Liu, T.-A.; Chen, S.-C.; Wang, J.; Jan, Y.-J.; Liou, J.-Y. Involvement of 14-3-3γ overexpression in extrahepatic metastasis of hepatocellular carcinoma. Hum. Pathol. 2011, 42, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-A.; Jan, Y.-J.; Ko, B.-S.; Wu, Y.-J.; Lu, Y.-J.; Liang, S.-M.; Liu, C.-C.; Chen, S.-C.; Wang, J.; Shyue, S.-K.; et al. Regulation of Aldo-keto-reductase family 1 B10 by 14-3-3ε and their prognostic impact of hepatocellular carcinoma. Oncotarget 2015, 6, 38967–38982. [Google Scholar] [CrossRef]
- Liu, T.-A.; Jan, Y.-J.; Ko, B.-S.; Liang, S.-M.; Chen, S.-C.; Wang, J.; Hsu, C.; Wu, Y.-M.; Liou, J.-Y. 14-3-3ε Overexpression Contributes to Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma. PLoS ONE 2013, 8, e57968. [Google Scholar] [CrossRef]
- Tang, S.; Bao, H.; Zhang, Y.; Yao, J.; Yang, P.; Chen, X. 14-3-3ε Mediates the Cell Fate Decision-Making Pathways in Response of Hepatocellular Carcinoma to Bleomycin-Induced DNA Damage. PLoS ONE 2013, 8, e55268. [Google Scholar] [CrossRef][Green Version]
- Holmes, T.R.; Al Matouq, J.; Holmes, M.; Sioda, N.; Rudd, J.C.; Bloom, C.; Nicola, L.; Palermo, N.Y.; Madson, J.G.; Lovas, S.; et al. Targeting 14-3-3ε activates apoptotic signaling to prevent cutaneous squamous cell carcinoma. Carcinogenesis 2020, 42, 232–242. [Google Scholar] [CrossRef]
- Liang, S.; Xu, Y.; Shen, G.; Liu, Q.; Zhao, X.; Xu, Z.; Xie, X.; Gong, F.; Li, R.; Wei, Y. Quantitative protein expression profiling of 14-3-3 isoforms in human renal carcinoma shows 14-3-3 epsilon is involved in limitedly increasing renal cell proliferation. Electrophoresis 2009, 30, 4152–4162. [Google Scholar] [CrossRef]
- Liou, J.-Y.; Ghelani, D.; Yeh, S.; Wu, K.K. Nonsteroidal Anti-inflammatory Drugs Induce Colorectal Cancer Cell Apoptosis by Suppressing 14-3-3epsilon. Cancer Res. 2007, 67, 3185–3191. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, J.; Ma, C.; Li, F.; Luo, H. Development and Validation of a Robust Ferroptosis-Related Prognostic Signature in Lung Adenocarcinoma. Front. Cell Dev. Biol. 2021, 9, 616271. [Google Scholar] [CrossRef]
- Jin, M.; Wu, L.; Chen, S.; Cai, R.; Dai, Y.; Yang, H.; Tang, L.; Li, Y. Arsenic trioxide enhances the chemotherapeutic efficiency of cisplatin in cholangiocarcinoma cells via inhibiting the 14-3-3ε-mediated survival mechanism. Cell Death Discov. 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Xue, Y.; Tang, S.; Yao, J.; Du, R.; Yang, P.; Chen, X. 14-3-3 Epsilon Dynamically Interacts with Key Components of Mitogen-Activated Protein Kinase Signal Module for Selective Modulation of the TNF-α-Induced Time Course-Dependent NF-κB Activity. J. Proteome Res. 2010, 9, 3465–3478. [Google Scholar] [CrossRef] [PubMed]
- He, Q.Q.; Wu, X.; Liu, X.P.; Yang, X.J.; Yuan, Z.M.; Zhang, Y. 14-3-3 epsilon plays an important role in testicular germ cell apoptosis: A functional proteomic study of experimental varicocele. Andrologia 2019, 51, e13275. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fulciniti, M.; Samur, M.K.; Ho, M.; Deng, S.; Liu, L.; Wen, K.; Yu, T.; Chyra, Z.; Dereibal, S.; et al. YWHAE/14-3-3ε expression impacts the protein load, contributing to proteasome inhibitor sensitivity in multiple myeloma. Blood 2020, 136, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Sai, Y.; Peng, K.; Ye, F.; Zhao, X.; Zhao, Y.; Zou, Z.; Cao, J.; Dong, Z. 14-3-3 Proteins in the Regulation of Rotenone-Induced Neurotoxicity Might be via Its Isoform 14-3-3Epsilon’s Involvement in Autophagy. Cell. Mol. Neurobiol. 2013, 33, 1109–1121. [Google Scholar] [CrossRef]
- Lee, S.K.; Park, S.O.; Joe, C.O.; Kim, Y.S. Interaction of HCV core protein with 14-3-3ε protein releases Bax to activate apoptosis. Biochem. Biophys. Res. Commun. 2007, 352, 756–762. [Google Scholar] [CrossRef]
- Tak, H.; Jang, E.; Kim, S.B.; Park, J.; Suk, J.; Yoon, Y.S.; Ahn, J.K.; Lee, J.-H.; Joe, C.O. 14-3-3epsilon inhibits MK5-mediated cell migration by disrupting F-actin polymerization. Cell. Signal. 2007, 19, 2379–2387. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, X.; Xu, H. miR-29b-3p inhibits 22Rv1 prostate cancer cell proliferation through the YWHAE/BCL-2 regulatory axis. Oncol. Lett. 2022, 24, 289. [Google Scholar] [CrossRef]
- Bjeije, H.; Soltani, B.M.; Behmanesh, M.; Zali, M.R. YWHAE long non-coding RNA competes with miR-323a-3p and miR-532-5p through activating K-Ras/Erk1/2 and PI3K/Akt signaling pathways in HCT116 cells. Hum. Mol. Genet. 2019, 28, 3219–3231. [Google Scholar] [CrossRef]
- Angeles, A.K.; Heckmann, D.; Flosdorf, N.; Duensing, S.; Sültmann, H. The ERG-Regulated LINC00920 Promotes Prostate Cancer Cell Survival via the 14-3-3ϵ–FOXO Pathway. Mol. Cancer Res. 2020, 18, 1545–1559. [Google Scholar] [CrossRef]
- Qi, W.; Liu, X.; Chen, W.; Li, Q.; Martinez, J.D. Overexpression of 14-3-3γ causes polyploidization in H322 lung cancer cells. Mol. Carcinog. 2007, 46, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Raungrut, P.; Wongkotsila, A.; Champoochana, N.; Lirdprapamongkol, K.; Svasti, J.; Thongsuksai, P. Knockdown of 14-3-3γ Suppresses Epithelial–Mesenchymal Transition and Reduces Metastatic Potential of Human Non-small Cell Lung Cancer Cells. Anticancer Res. 2018, 38, 3507–3514. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Y.; Dai, D.-F.; Hua, Y.; Qi, W.-Q. p53 suppresses 14-3-3γ by stimulating proteasome-mediated 14-3-3γ protein degradation. Int. J. Oncol. 2014, 46, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-O.; Kim, S.-R.; Lim, K.-H.; Kim, J.-H.; Ajjappala, B.; Lee, H.-J.; Choi, J.-I.; Baek, K.-H. Deubiquitinating enzyme USP37 regulating oncogenic function of 14-3-3γ. Oncotarget 2015, 6, 36551–36576. [Google Scholar] [CrossRef]
- Gomes, C.J.; Centuori, S.M.; Harman, M.W.; Putnam, C.W.; Wolgemuth, C.W.; Martinez, J.D. The induction of endoreduplication and polyploidy by elevated expression of 14-3-3γ. Genes Cancer 2017, 8, 771–783. [Google Scholar] [CrossRef][Green Version]
- Lee, Y.-S.; Lee, J.K.; Bae, Y.; Lee, B.-S.; Kim, E.; Cho, C.-H.; Ryoo, K.; Yoo, J.; Kim, C.-H.; Yi, G.-S.; et al. Suppression of 14-3-3γ-mediated surface expression of ANO1 inhibits cancer progression of glioblastoma cells. Sci. Rep. 2016, 6, 26413. [Google Scholar] [CrossRef]
- Wu, Z.; Weng, D.; Li, G. Quantitative proteome analysis of overexpressed Cripto-1 tumor cell reveals 14-3-3γ as a novel biomarker in nasopharyngeal carcinoma. J. Proteom. 2013, 83, 26–36. [Google Scholar] [CrossRef]
- Song, Y.; Yang, Z.; Ke, Z.; Yao, Y.; Hu, X.; Sun, Y.; Li, H.; Yin, J.; Zeng, C. Expression of 14-3-3γ in patients with breast cancer: Correlation with clinicopathological features and prognosis. Cancer Epidemiol. 2012, 36, 533–536. [Google Scholar] [CrossRef]
- Zhao, Y.; Yue, S.; Zhou, X.; Guo, J.; Ma, S.; Chen, Q. O-GlcNAc transferase promotes the nuclear localization of the focal adhesion–associated protein Zyxin to regulate UV-induced cell death. J. Biol. Chem. 2022, 298, 101776. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Sehgal, L.; Bose, A.; Gulvady, A.; Senapati, P.; Thorat, R.; Basu, S.; Bhatt, K.; Hosing, A.S.; Balyan, R.; et al. 14-3-3γ Prevents Centrosome Amplification and Neoplastic Progression. Sci. Rep. 2016, 6, 26580. [Google Scholar] [CrossRef]
- Tilwani, S.; Gandhi, K.; Narayan, S.; Ainavarapu, S.R.K.; Dalal, S.N. Disruption of desmosome function leads to increased centrosome clustering in 14-3-3γ-knockout cells with supernumerary centrosomes. FEBS Lett. 2021, 595, 2675–2690. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qiao, W.; Wang, X.; Yin, H.; Cui, J.; Cui, Y.; Chen, X.; Hu, J.; Lu, H.; Meng, Q.; et al. 14-3-3ζ/TGFβR1 promotes tumor metastasis in lung squamous cell carcinoma. Oncotarget 2016, 7, 82972–82984. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Kong, L.; Liang, K.; Wu, Y.; Jin, H.; Song, B.; Tan, X. Identification of dissociation factors in pancreatic Cancer using a mass spectrometry-based proteomic approach. BMC Cancer 2020, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Hu, X.; Zhou, L.; Zou, S.; Sun, L.-Z.; Zhu, X. Overexpression of the 14-3-3γ protein in uterine leiomyoma cells results in growth retardation and increased apoptosis. Cell. Signal. 2018, 45, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, E.; Mimae, T.; Ito, M.; Kadoya, T.; Miyata, Y.; Ito, A.; Okada, M. Breast cancer cell motility is promoted by 14-3-3γ. Breast Cancer 2019, 26, 581–593. [Google Scholar] [CrossRef]
- Mariani, R.A.; Paranjpe, S.; Dobrowolski, R.; Weber, G.F. 14-3-3 targets keratin intermediate filaments to mechanically sensitive cell–cell contacts. Mol. Biol. Cell 2020, 31, 930–943. [Google Scholar] [CrossRef]
- Teo, Z.; Sng, M.K.; Chan, J.S.K.; Lim, M.M.K.; Li, Y.; Li, L.; Phua, T.; Lee, J.Y.H.; Tan, Z.W.; Zhu, P.; et al. Elevation of adenylate energy charge by angiopoietin-like 4 enhances epithelial–mesenchymal transition by inducing 14-3-3γ expression. Oncogene 2017, 36, 6408–6419. [Google Scholar] [CrossRef]
- Kasahara, K.; Goto, H.; Izawa, I.; Kiyono, T.; Watanabe, N.; Elowe, S.; A Nigg, E.; Inagaki, M. PI 3-kinase-dependent phosphorylation of Plk1–Ser99 promotes association with 14-3-3γ and is required for metaphase–anaphase transition. Nat. Commun. 2013, 4, 1882. [Google Scholar] [CrossRef]
- Lee, J.-H.; Lu, H. 14-3-3γ Inhibition of MDMX-mediated p21 Turnover Independent of p53. J. Biol. Chem. 2011, 286, 5136–5142. [Google Scholar] [CrossRef]
- Xiong, X.X.; Hu, D.X.; Xu, L.; Lin, H.; Zhang, Y.; Li, C.Y.; Chen, X.Q. Selective 14-3-3γ Upregulation Promotes Beclin-1-LC3-Autophagic Influx via β-Catenin Interaction in Starved Neurons In Vitro and In Vivo. Neurochem. Res. 2019, 44, 849–858. [Google Scholar] [CrossRef]
- Radhakrishnan, V.M.; Martinez, J.D. 14-3-3γ Induces Oncogenic Transformation by Stimulating MAP Kinase and PI3K Signaling. PLoS ONE 2010, 5, e11433. [Google Scholar] [CrossRef] [PubMed]
- Ajjappala, B.S.; Kim, Y.-S.; Kim, M.-S.; Lee, M.-Y.; Lee, K.-Y.; Ki, H.-Y.; Cha, D.H.; Baek, K.-H. 14-3-3γ Is Stimulated by IL-3 and Promotes Cell Proliferation. J. Immunol. 2009, 182, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Hosing, A.S.; Kundu, S.; Dalal, S.N. 14-3-3 gamma is required to enforce both the incomplete S phase and G2 DNA damage checkpoints. Cell Cycle 2008, 7, 3171–3179. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Q.; Chen, J.G.; Zhang, Y.; Hsiao, W.W.L.; Yu, A.C.H. 14-3-3γ Is upregulated by in vitro ischemia and binds to protein kinase Raf in primary cultures of astrocytes. Glia 2003, 42, 315–324. [Google Scholar] [CrossRef]
- Wang, P.; Deng, Y.; Fu, X. MiR-509-5p suppresses the proliferation, migration, and invasion of non-small cell lung cancer by targeting YWHAG. Biochem. Biophys. Res. Commun. 2017, 482, 935–941. [Google Scholar] [CrossRef]
- Chu, Y.-W.; Wang, C.-R.; Weng, F.-B.; Yan, Z.-J.; Wang, C. MicroRNA-222 contributed to cell proliferation, invasion and migration via regulating YWHAG in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2588–2597. [Google Scholar]
- Bai, Q.; Yu, J.; Li, Y.; Ma, J.; Gou, Y. MicroRNA-182 promoted esophageal squamous cell carcinoma cell growth and metastasis via targeting YWHAG. J. BUON 2018, 23, 1439–1447. [Google Scholar]
- Ni, J.; Wang, J.; Fu, Y.; Yan, C.; Zhu, M.; Jiang, Y.; Chen, J.; Ding, Y.; Fan, X.; Li, G.; et al. Functional genetic variants in centrosome-related genes CEP72 and YWHAG confer susceptibility to gastric cancer. Arch. Toxicol. 2020, 94, 2861–2872. [Google Scholar] [CrossRef]
- Xu, J.; Wang, J.; He, Z.; Chen, P.; Jiang, X.; Chen, Y.; Liu, X.; Jiang, J. LncRNA CERS6-AS1 promotes proliferation and metastasis through the upregulation of YWHAG and activation of ERK signaling in pancreatic cancer. Cell Death Dis. 2021, 12, 648. [Google Scholar] [CrossRef]
- Zheng, Y.; Miao, Y.; Xie, J.; Lin, Y.; Yao, Q.; Cai, J.; Yang, X. Long noncoding RNA lysophospholipase-like 1-2 as ceRNA modulates glioma metastasis by regulating miR-217/YWHAG. Am. J. Transl. Res. 2020, 12, 4204–4215. [Google Scholar]
- Wang, H.; Zhi, H.; Ma, D.; Li, T. MiR-217 promoted the proliferation and invasion of glioblastoma by repressing YWHAG. Cytokine 2017, 92, 93–102. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, Z.; Li, Y.; Wang, L.; Liu, Y. LncRNA PTPRG-AS1 Promotes the Metastasis of Hepatocellular Carcinoma by Enhancing YWHAG. J. Oncol. 2021, 2021, 3624306. [Google Scholar] [CrossRef] [PubMed]
- Bhawal, U.K.; Sugiyama, M.; Nomura, Y.; Kuniyasu, H.; Tsukinoki, K. Loss of 14-3-3 Sigma Protein Expression and Presence of Human Papillomavirus Type 16 E6 in Oral Squamous Cell Carcinoma. Arch. Otolaryngol.—Head Neck Surg. 2008, 134, 1055–1059. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Simooka, H.; Oyama, T.; Sano, T.; Horiguchi, J.; Nakajima, T. Immunohistochemical analysis of 14-3-3 sigma and related proteins in hyperplastic and neoplastic breast lesions, with particular reference to early carcinogenesis. Pathol. Int. 2004, 54, 595–602. [Google Scholar] [CrossRef]
- Qi, Y.-J.; Wang, M.; Liu, R.-M.; Wei, H.; Chao, W.-X.; Zhang, T.; Lou, Q.; Li, X.-M.; Ma, J.; Zhu, H.; et al. Downregulation of 14-3-3σ Correlates with Multistage Carcinogenesis and Poor Prognosis of Esophageal Squamous Cell Carcinoma. PLoS ONE 2014, 9, e95386. [Google Scholar] [CrossRef]
- Schultz, J.; Ibrahim, S.M.; Vera, J.; Kunz, M. 14-3-3σ gene silencing during melanoma progression and its role in cell cycle control and cellular senescence. Mol. Cancer 2009, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H.; Lengauer, C.; Polyak, K.; He, T.-C.; Zhang, L.; Thiagalingam, S.; Kinzler, K.W.; Vogelstein, B. 14-3-3σIs a p53-Regulated Inhibitor of G2/M Progression. Mol. Cell 1997, 1, 3–11. [Google Scholar] [CrossRef]
- Laronga, C.; Yang, H.-Y.; Neal, C.; Lee, M.-H. Association of the Cyclin-dependent Kinases and 14-3-3 Sigma Negatively Regulates Cell Cycle Progression. J. Biol. Chem. 2000, 275, 23106–23112. [Google Scholar] [CrossRef]
- Han, Z.; Dimas, K.; Tian, X.; Wang, Y.; Hemmi, H.; Yamada, K.; Kato, N.; Pantazis, P.; Ramanujam, R.J.; Anant, S.; et al. 14-3-3sigma-dependent resistance to cisplatin. Anticancer Res. 2009, 29, 2009–2014. [Google Scholar]
- Sirivatanauksorn, V.; Dumronggittigule, W.; Dulnee, B.; Srisawat, C.; Sirivatanauksorn, Y.; Pongpaibul, A.; Masaratana, P.; Somboonyosdech, C.; Sripinitchai, S.; Kositamongkol, P.; et al. Role of stratifin (14-3-3 sigma) in adenocarcinoma of gallbladder: A novel prognostic biomarker. Surg. Oncol. 2019, 32, 57–62. [Google Scholar] [CrossRef]
- Cheng, L.; Pan, C.-X.; Zhang, J.-T.; Zhang, S.; Kinch, M.S.; Li, L.; Baldridge, L.A.; Wade, C.; Hu, Z.; Koch, M.O.; et al. Loss of 14-3-3σ in Prostate Cancer and Its Precursors. Clin. Cancer Res. 2004, 10, 3064–3068. [Google Scholar] [CrossRef] [PubMed]
- Holm, R.; Ali, T.; Svendsrud, D.H.; Nesland, J.M.; Kristensen, G.B.; Lyng, H. Expression of 14-3-3σ in cervical squamous cell carcinomas: Relationship with clinical outcome. Oncol. Rep. 2009, 22, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Hustinx, S.R.; Fukushima, N.; Zahurak, M.L.; Riall, T.S.; Maitra, A.; Brosens, L.; Cameron, J.L.; Yeo, C.J.; Offerhaus, G.J.A.; Hruban, R.H.; et al. Expression and prognostic significance of 14-3-3 sigma and ERM family protein expression in periampullary neoplasms. Cancer Biol. Ther. 2005, 4, 596–601. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanaka, K.; Hatada, T.; Kobayashi, M.; Mohri, Y.; Tonouchi, H.; Miki, C.; Nobori, T.; Kusunoki, M. The clinical implication of 14-3-3 sigma expression in primary gastrointestinal malignancy. Int. J. Oncol. 2004, 25, 1591–1597. [Google Scholar] [CrossRef] [PubMed]
- Mhawech, P.; Greloz, V.; Assaly, M.; Herrmann, F. Immunohistochemical expression of 14-3-3 sigma protein in human urological and gynecological tumors using a multi-tumor microarray analysis. Pathol. Int. 2005, 55, 77–82. [Google Scholar] [CrossRef]
- Ghaffari, A.; Li, Y.; Kilani, R.T.; Ghahary, A. 14-3-3σ associates with cell surface aminopeptidase N in the regulation of matrix metalloproteinase-1. J. Cell Sci. 2010, 123, 2996–3005. [Google Scholar] [CrossRef][Green Version]
- Mühlmann, G.; Öfner, D.; Zitt, M.; Müller, H.M.; Maier, H.; Moser, P.; Schmid, K.W.; Zitt, M.; Amberger, A. 14-3-3 sigma and p53 expression in gastric cancer and its clinical applications. Dis. Markers 2010, 29, 21–29. [Google Scholar] [CrossRef]
- Kim, Y.; Shiba-Ishii, A.; Nakagawa, T.; Iemura, S.-I.; Natsume, T.; Nakano, N.; Matsuoka, R.; Sakashita, S.; Lee, S.; Kawaguchi, A.; et al. Stratifin regulates stabilization of receptor tyrosine kinases via interaction with ubiquitin-specific protease 8 in lung adenocarcinoma. Oncogene 2018, 37, 5387–5402. [Google Scholar] [CrossRef]
- Sun, N.; Wu, Y.; Huang, B.; Liu, Q.; Dong, Y.; Ding, J.; Liu, Y. Decreased expression of 14-3-3 σ, an early event of malignant transformation of respiratory epithelium, also facilitates progression of squamous cell lung cancer. Thorac. Cancer 2015, 6, 715–721. [Google Scholar] [CrossRef]
- Ma, Y.-S.; Hou, L.-K.; Yao, S.-H.; Liu, J.-B.; Yu, X.-C.; Shi, Y.; Yang, X.-L.; Wu, W.; Wu, C.-Y.; Jiang, G.-X.; et al. Elevated Stratifin promotes cisplatin-based chemotherapy failure and poor prognosis in non-small cell lung cancer. Mol. Ther.-Oncolytics 2021, 22, 326–335. [Google Scholar] [CrossRef]
- Pulukuri, S.M.; Rao, J.S. CpG island promoter methylation and silencing of 14-3-3σ gene expression in LNCaP and Tramp-C1 prostate cancer cell lines is associated with methyl-CpG-binding protein MBD2. Oncogene 2006, 25, 4559–4572. [Google Scholar] [CrossRef] [PubMed]
- Kunze, E.; Wendt, M.; Schlott, T. Promoter hypermethylation of the 14-3-3 σ, SYK and CAGE-1 genes is related to the various phenotypes of urinary bladder carcinomas and associated with progression of transitional cell carcinomas. Int. J. Mol. Med. 2006, 18, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.; Tan, S.-X.; Tang, C.-E.; Huang, W.-G.; Cheng, A.-L.; Li, C.; Zhang, P.-F.; Li, M.-Y.; Li, J.-L.; Yi, H.; et al. Inactivation of 14-3-3 σ by promoter methylation correlates with metastasis in nasopharyngeal carcinoma. J. Cell. Biochem. 2009, 106, 858–866. [Google Scholar] [CrossRef]
- Ye, M.; Huang, T.; Ying, Y.; Li, J.; Yang, P.; Ni, C.; Zhou, C.; Chen, S. Detection of 14-3-3 sigma (σ) promoter methylation as a noninvasive biomarker using blood samples for breast cancer diagnosis. Oncotarget 2016, 8, 9230–9242. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Kim, J.Y.; Jeong, J.; Lee, J.E.; Yang, W.I.; Jung, W.H. The Role and Regulatory Mechanism of 14-3-3 Sigma in Human Breast Cancer. J. Breast Cancer 2014, 17, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Shiba-Ishii, A. Significance of stratifin in early progression of lung adenocarcinoma and its potential therapeutic relevance. Pathol. Int. 2021, 71, 655–665. [Google Scholar] [CrossRef]
- Raungrut, P.; Petjaroen, P.; Geater, S.L.; Keeratichananont, W.; Phukaoloun, M.; Suwiwat, S.; Thongsuksai, P. Methylation of 14-3-3σ gene and prognostic significance of 14-3-3σ expression in non-small cell lung cancer. Oncol. Lett. 2017, 14, 5257–5264. [Google Scholar] [CrossRef]
- Mhawech, P.; Benz, A.; Cerato, C.; Greloz, V.; Assaly, M.; Desmond, J.C.; Koeffler, H.P.; Lodygin, D.; Hermeking, H.; Herrmann, F.; et al. Downregulation of 14-3-3σ in ovary, prostate and endometrial carcinomas is associated with CpG island methylation. Mod. Pathol. 2004, 18, 340–348. [Google Scholar] [CrossRef]
- Uchida, D.; Begum, N.-M.; Almofti, A.; Kawamata, H.; Yoshida, H.; Sato, M. Frequent downregulation of 14-3-3 σ protein and hypermethylation of 14-3-3 σ gene in salivary gland adenoid cystic carcinoma. Br. J. Cancer 2004, 91, 1131–1138. [Google Scholar] [CrossRef][Green Version]
- Akahira, J.-I.; Sugihashi, Y.; Suzuki, T.; Ito, K.; Niikura, H.; Moriya, T.; Nitta, M.; Okamura, H.; Inoue, S.; Sasano, H.; et al. Decreased Expression of 14-3-3σ Is Associated with Advanced Disease in Human Epithelial Ovarian Cancer: Its correlation with aberrant DNA methylation. Clin. Cancer Res. 2004, 10, 2687–2693. [Google Scholar] [CrossRef]
- Gasco, M.; Bell, A.K.; Heath, V.; Sullivan, A.; Smith, P.; Hiller, L.; Yulug, I.; Numico, G.; Merlano, M.; Farrell, P.J.; et al. Epigenetic inactivation of 14-3-3 sigma in oral carcinoma: Association with p16(INK4a) silencing and human papillomavirus negativity. Cancer Res. 2002, 62, 2072–2076. [Google Scholar] [PubMed]
- Sano, T.; Shimooka, H.; Weixa, P.; Segawa, A.; Jian, Z.; Motegi, A.; Nakayama, H.; Oyama, T.; Nakajima, T. Immunohistochemical expression of 14-3-3 sigma protein in various histological subtypes of uterine cervical cancers. Pathol. Int. 2004, 54, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-J.; Deng, G.; Xiao, Z.-Q.; Yao, H.-X.; Li, C.; Peng, F.; Li, M.-Y.; Zhang, P.-F.; Chen, Y.-H.; Chen, Z.-C. Identificating 14-3-3 sigma as a lymph node metastasis-related protein in human lung squamous carcinoma. Cancer Lett. 2009, 279, 65–73. [Google Scholar] [CrossRef]
- Khongmanee, A.; Lirdprapamongkol, K.; Tit-Oon, P.; Chokchaichamnankit, D.; Svasti, J.; Srisomsap, C. Proteomic analysis reveals important role of 14-3-3σ in anoikis resistance of cholangiocarcinoma cells. Proteomics 2013, 13, 3157–3166. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wen, Y.-Y.; Zhao, R.; Lin, Y.-L.; Fournier, K.; Yang, H.-Y.; Qiu, Y.; Diaz, J.; Laronga, C.; Lee, M.-H. DNA Damage–Induced Protein 14-3-3 σ Inhibits Protein Kinase B/Akt Activation and Suppresses Akt-Activated Cancer. Cancer Res. 2006, 66, 3096–3105. [Google Scholar] [CrossRef] [PubMed]
- Moreira, J.M.A.; Gromov, P.; Celis, J.E. Expression of the Tumor Suppressor Protein 14-3-3σ Is Down-regulated in Invasive Transitional Cell Carcinomas of the Urinary Bladder Undergoing Epithelial-to-Mesenchymal Transition. Mol. Cell. Proteom. 2004, 3, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-B.; Zhao, Y.-J.; Wang, X.-Y.; Liao, W.-J.; Chen, P.; Deng, K.-J.; Cong, X.; Fei, R.; Wu, X.; Shao, Q.-X.; et al. Regulatory factor X5 promotes hepatocellular carcinoma progression by transactivating tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta and suppressing apoptosis. Chin. Med. J. 2019, 132, 1572–1581. [Google Scholar] [CrossRef]
- Singrang, N.; Kittisenachai, S.; Roytrakul, S.; Svasti, J.; Kangsamaksin, T. NOTCH1 regulates the viability of cholangiocarcinoma cells via 14-3-3 theta. J. Cell Commun. Signal. 2018, 13, 245–254. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Z.; Zhu, Z.; Liu, J.; Zheng, X.; Zhang, Y. Network analysis of ChIP-Seq data reveals key genes in prostate cancer. Eur. J. Med. Res. 2014, 19, 47. [Google Scholar] [CrossRef]
- Sanchez, R.; St-Cyr, J.; LaLonde, M.-E.; Healy, J.; Richer, C.; Gagné, V.; Laverdière, C.; Silverman, L.B.; Sallan, S.E.; Neuberg, D.; et al. Impact of promoter polymorphisms in key regulators of the intrinsic apoptosis pathway on the outcome of childhood acute lymphoblastic leukemia. Haematologica 2013, 99, 314–321. [Google Scholar] [CrossRef]
- Hou, H.; Lyu, Y.; Jiang, J.; Wang, M.; Zhang, R.; Liew, C.-C.; Wang, B.; Cheng, C. Peripheral blood transcriptome identifies high-risk benign and malignant breast lesions. PLoS ONE 2020, 15, e0233713. [Google Scholar] [CrossRef]
- Qiu, J.; Choi, G.; Li, L.; Wang, H.; Pitteri, S.J.; Pereira-Faca, S.R.; Krasnoselsky, A.L.; Randolph, T.W.; Omenn, G.S.; Edelstein, C.; et al. Occurrence of Autoantibodies to Annexin I, 14-3-3 Theta and LAMR1 in Prediagnostic Lung Cancer Sera. J. Clin. Oncol. 2008, 26, 5060–5066. [Google Scholar] [CrossRef]
- Pereira-Faca, S.R.; Kuick, R.; Puravs, E.; Zhang, Q.; Krasnoselsky, A.L.; Phanstiel, D.; Qiu, J.; Misek, D.E.; Hinderer, R.; Tammemagi, M.; et al. Identification of 14-3-3θ as an Antigen that Induces a Humoral Response in Lung Cancer. Cancer Res. 2007, 67, 12000–12006. [Google Scholar] [CrossRef][Green Version]
- Nomura, M.; Shimizu, S.; Sugiyama, T.; Narita, M.; Ito, T.; Matsuda, H.; Tsujimoto, Y. 14-3-3 Interacts Directly with and Negatively Regulates Pro-apoptotic Bax. J. Biol. Chem. 2003, 278, 2058–2065. [Google Scholar] [CrossRef]
- Schuster, T.B.; Costina, V.; Findeisen, P.; Neumaier, M.; Ahmad-Nejad, P. Identification and Functional Characterization of 14-3-3 in TLR2 Signaling. J. Proteome Res. 2011, 10, 4661–4670. [Google Scholar] [CrossRef]
- Nakamura, T.; Hayashi, T.; Mimori-Kiyosue, Y.; Sakaue, F.; Matsuura, K.; Iemura, S.-I.; Natsume, T.; Akiyama, T. The PX-RICS-14-3-3ζ/θ Complex Couples N-cadherin-β-Catenin with Dynein-Dynactin to Mediate Its Export from the Endoplasmic Reticulum. J. Biol. Chem. 2010, 285, 16145–16154. [Google Scholar] [CrossRef]
- Pierrat, B.; Ito, M.; Hinz, W.; Simonen, M.; Erdmann, D.; Chiesi, M.; Heim, J. Uncoupling proteins 2 and 3 interact with members of the 14.3.3 family. Eur. J. Biochem. 2000, 267, 2680–2687. [Google Scholar] [CrossRef]
- Yan, Y.; Xu, Y.; Gao, Y.-Y.; Zong, Z.-H.; Zhang, Q.; Li, C.; Wang, H.-Q. Implication of 14-3-3ε and 14-3-3θ/τ in proteasome inhibition-induced apoptosis of glioma cells. Cancer Sci. 2012, 104, 55–61. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, X.; Guan, Y.; Gong, M.; He, K.; Huang, B. 1,3-Dicaffeoylquinic acid targeting 14-3-3 tau suppresses human breast cancer cell proliferation and metastasis through IL6/JAK2/PI3K pathway. Biochem. Pharmacol. 2020, 172, 113752. [Google Scholar] [CrossRef]
- Vazquez, A.; Grochola, L.F.; Bond, E.E.; Levine, A.J.; Taubert, H.; Müller, T.H.; Würl, P.; Bond, G.L. Chemosensitivity Profiles Identify Polymorphisms in the p53 Network Genes 14-3-3τ and CD44 That Affect Sarcoma Incidence and Survival. Cancer Res. 2010, 70, 172–180. [Google Scholar] [CrossRef]
- Li, J.; Xu, H.; Wang, Q.; Wang, S.; Xiong, N. 14-3-3ζ promotes gliomas cells invasion by regulating Snail through the PI3K/AKT signaling. Cancer Med. 2019, 8, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Rajendra, J.; Ghorai, A.; Dutt, S. 14-3-3ζ negatively regulates mitochondrial biogenesis in GBM residual cells. Heliyon 2021, 7, e08371. [Google Scholar] [CrossRef]
- Cao, W.-D.; Kawai, N.; Miyake, K.; Zhang, X.; Fei, Z.; Tamiya, T. Relationship of 14-3-3zeta (ζ), HIF-1α, and VEGF expression in human brain gliomas. Brain Tumor Pathol. 2013, 31, 1–10. [Google Scholar] [CrossRef]
- Luo, Z.-J.; Zhang, X.-F.; Rapp, U.; Avruch, J. Identification of the 14.3.3 ζ Domains Important for Self-association and Raf Binding. J. Biol. Chem. 1995, 270, 23681–23687. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Y.; He, J.-C.; Wang, J.-M.; Schemmer, P.; Ma, C.-Q.; Qian, Y.-W.; Yao, W.; Zhang, J.; Qi, W.-P.; et al. 14-3-3ζ and aPKC-ι synergistically facilitate epithelial-mesenchymal transition of cholangiocarcinoma via GSK-3β/snail signaling pathway. Oncotarget 2016, 7, 55191–55210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, D.; Yi, B.; Liao, Z.; Tang, L.; Yin, D.; Zeng, S.; Yao, J.; He, M. 14-3-3γ protein attenuates lipopolysaccharide-induced cardiomyocytes injury through the Bcl-2 family/mitochondria pathway. Int. Immunopharmacol. 2014, 21, 509–515. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Ke, A.-W.; Shi, G.-M.; Zhang, X.; Zhang, C.; Shi, Y.-H.; Wang, X.-Y.; Ding, Z.-B.; Xiao, Y.-S.; Yan, J.; et al. αB-crystallin complexes with 14-3-3ζ to induce epithelial-mesenchymal transition and resistance to sorafenib in hepatocellular carcinoma. Hepatology 2013, 57, 2235–2247. [Google Scholar] [CrossRef]
- Wang, X.; Shen, H.; Zhangyuan, G.; Huang, R.; Zhang, W.; He, Q.; Jin, K.; Zhuo, H.; Zhang, Z.; Wang, J.; et al. 14-3-3ζ delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. 2018, 9, 159. [Google Scholar] [CrossRef]
- Zhao, J.; Fu, X.; Chen, H.; Min, L.; Sun, J.; Yin, J.; Guo, J.; Li, H.; Tang, Z.; Ruan, Y.; et al. G3BP1 interacts with YWHAZ to regulate chemoresistance and predict adjuvant chemotherapy benefit in gastric cancer. Br. J. Cancer 2020, 124, 425–436. [Google Scholar] [CrossRef]
- Nishimura, Y.; Komatsu, S.; Ichikawa, D.; Nagata, H.; Hirajima, S.; Takeshita, H.; Kawaguchi, T.; Arita, T.; Konishi, H.; Kashimoto, K.; et al. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br. J. Cancer 2013, 108, 1324–1331. [Google Scholar] [CrossRef]
- Watanabe, N.; Komatsu, S.; Ichikawa, D.; Miyamae, M.; Ohashi, T.; Okajima, W.; Kosuga, T.; Konishi, H.; Shiozaki, A.; Fujiwara, H.; et al. Overexpression of YWHAZ as an independent prognostic factor in adenocarcinoma of the esophago-gastric junction. Am. J. Cancer Res. 2016, 6, 2729–2736. [Google Scholar]
- Liu, M.; Liu, X.; Ren, P.; Li, J.; Chai, Y.; Zheng, S.-J.; Chen, Y.; Duan, Z.-P.; Li, N.; Zhang, J.-Y. A cancer-related protein 14-3-3ζ is a potential tumor-associated antigen in immunodiagnosis of hepatocellular carcinoma. Tumor Biol. 2014, 35, 4247–4256. [Google Scholar] [CrossRef][Green Version]
- Zhang, C.; Liu, L.-X.; Dong, Z.-R.; Shi, G.-M.; Cai, J.-B.; Zhang, P.-F.; Ke, A.-W.; Yu, J.-X.; Zhou, J.; Fan, J. Up-regulation of 14-3-3ζ expression in intrahepatic cholangiocarcinoma and its clinical implications. Tumor Biol. 2014, 36, 1781–1789. [Google Scholar] [CrossRef]
- Masui, O.; White, N.M.A.; DeSouza, L.V.; Krakovska, O.; Matta, A.; Metias, S.; Khalil, B.; Romaschin, A.D.; Honey, R.J.; Stewart, R.; et al. Quantitative Proteomic Analysis in Metastatic Renal Cell Carcinoma Reveals a Unique Set of Proteins with Potential Prognostic Significance. Mol. Cell. Proteom. 2013, 12, 132–144. [Google Scholar] [CrossRef]
- Chen, M.; Huang, H.; He, H.; Ying, W.; Liu, X.; Dai, Z.; Yin, J.; Mao, N.; Qian, X.; Pan, L. Quantitative proteomic analysis of mitochondria from human ovarian cancer cells and their paclitaxel-resistant sublines. Cancer Sci. 2015, 106, 1075–1083. [Google Scholar] [CrossRef]
- Yu, C.; Li, C.; Chen, I.; Lai, M.; Lin, Z.; Korla, P.K.; Chai, C.; Ko, G.; Chen, C.; Hwang, T.; et al. YWHAZ amplification/overexpression defines aggressive bladder cancer and contributes to chemo-/radio-resistance by suppressing caspase-mediated apoptosis. J. Pathol. 2019, 248, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Takayama, K.-I.; Urano, T.; Fujimura, T.; Ashikari, D.; Obinata, D.; Horie-Inoue, K.; Takahashi, S.; Ouchi, Y.; Homma, Y.; et al. 14-3-3ζ, a Novel Androgen-Responsive Gene, Is Upregulated in Prostate Cancer and Promotes Prostate Cancer Cell Proliferation and Survival. Clin. Cancer Res. 2012, 18, 5617–5627. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, X.; Liao, Z.; Liang, H.; Chu, L.; Dong, W.; Zhang, X.; Ge, Q.; Liu, Q.; Fan, P.; et al. 14–3-3ζ inhibits heme oxygenase-1 (HO-1) degradation and promotes hepatocellular carcinoma proliferation: Involvement of STAT3 signaling. J. Exp. Clin. Cancer Res. 2019, 38, 3. [Google Scholar] [CrossRef]
- Bin Seo, S.; Baek, J.-Y.; Lim, J.-H.; Jin, X.; Lee, M.-Y.; Lee, J.-H. 14-3-3ζ targeting induced senescence in Hep-2 laryngeal cancer cell through deneddylation of Cullin1 in the Skp1-Cullin-F-box protein complex. Cell Prolif. 2019, 52, e12654. [Google Scholar] [CrossRef]
- Lin, M.; Morrison, C.D.; Jones, S.; Mohamed, N.; Bacher, J.; Plass, C. Copy number gain and oncogenic activity of YWHAZ/14-3-3ζ in head and neck squamous cell carcinoma. Int. J. Cancer 2009, 125, 603–611. [Google Scholar] [CrossRef]
- Fan, T.; Li, R.; Todd, N.W.; Qiu, Q.; Fang, H.-B.; Wang, H.; Shen, J.; Zhao, R.Y.; Caraway, N.P.; Katz, R.L.; et al. Up-regulation of 14-3-3ζ in Lung Cancer and Its Implication as Prognostic and Therapeutic Target. Cancer Res. 2007, 67, 7901–7906. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.-Y.; Ding, J.-Y.; Gu, J.; Lu, C.-L.; Lin, Z.-W.; Guo, J.; Ge, D. The overexpression of 14-3-3ζ and Hsp27 promotes non-small cell lung cancer progression. Cancer 2013, 120, 652–663. [Google Scholar] [CrossRef]
- Jin, L.M.; Han, X.H.; Jie, Y.Q.; Meng, S.S. 14-3-3ζ silencing retards tongue squamous cell carcinoma progression by inhibiting cell survival and migration. Cancer Gene Ther. 2016, 23, 206–213. [Google Scholar] [CrossRef]
- Yang, X.; Cao, W.; Wang, X.; Zhang, X.; Zhang, W.; Li, Z.; Fu, H. Down-regulation of 14-3-3zeta reduces proliferation and increases apoptosis in human glioblastoma. Cancer Gene Ther. 2019, 27, 399–411. [Google Scholar] [CrossRef]
- Shi, J.; Ye, J.; Fei, H.; Jiang, S.; Wu, Z.; Chen, Y.; Zhang, L.; Yang, X. YWHAZ promotes ovarian cancer metastasis by modulating glycolysis. Oncol. Rep. 2018, 41, 1101–1112. [Google Scholar] [CrossRef]
- Tong, S.; Chen, S.-C.; Xu, K.-Y.; Fang, B.; Wang, S.-H.; Wang, J.-J. 14-3-3ζ promotes esophageal squamous cell carcinoma invasion by repressing S1PR2 protein expression through NF-κB signaling. Arch. Biochem. Biophys. 2018, 643, 7–13. [Google Scholar] [CrossRef]
- Bergamaschi, A.; Frasor, J.; Borgen, K.; Stanculescu, A.; Johnson, P.; Rowland, K.; Wiley, E.L.; Katzenellenbogen, B.S. 14-3-3ζ as a predictor of early time to recurrence and distant metastasis in hormone receptor-positive and -negative breast cancers. Breast Cancer Res. Treat. 2012, 137, 689–696. [Google Scholar] [CrossRef]
- Neal, C.L.; Xu, J.; Li, P.; Mori, S.; Yang, J.; Neal, N.N.; Zhou, X.; Wyszomierski, S.L.; Yu, D. Overexpression of 14-3-3ζ in cancer cells activates PI3K via binding the p85 regulatory subunit. Oncogene 2011, 31, 897–906. [Google Scholar] [CrossRef]
- Lu, J.; Guo, H.; Treekitkarnmongkol, W.; Li, P.; Zhang, J.; Shi, B.; Ling, C.; Zhou, X.; Chen, T.; Chiao, P.J.; et al. 14-3-3ζ Cooperates with ErbB2 to Promote Ductal Carcinoma In Situ Progression to Invasive Breast Cancer by Inducing Epithelial-Mesenchymal Transition. Cancer Cell 2009, 16, 195–207. [Google Scholar] [CrossRef]
- Kambach, D.M.; Sodi, V.L.; I Lelkes, P.; Azizkhan-Clifford, J.; Reginato, M.J. ErbB2, FoxM1 and 14-3-3ζ prime breast cancer cells for invasion in response to ionizing radiation. Oncogene 2013, 33, 589–598. [Google Scholar] [CrossRef]
- Kim, H.-J.; Sung, S.H.; Kim, C.Y.; Bae, M.K.; Cho, M.S.; Kim, Y.H.; Kim, S.C.; Ju, W. 14-3-3ζ Overexpression is Associated with Poor Prognosis in Ovarian Cancer. Yonsei Med. J. 2018, 59, 51–56. [Google Scholar] [CrossRef]
- Rüenauver, K.; Menon, R.; A Svensson, M.; Carlsson, J.; Vogel, W.; Andrén, O.; Nowak, M.; Perner, S. Prognostic significance of YWHAZ expression in localized prostate cancer. Prostate Cancer Prostatic Dis. 2014, 17, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Goc, A.; Abdalla, M.; Al-Azayzih, A.; Somanath, P.R. Rac1 Activation Driven by 14-3-3ζ Dimerization Promotes Prostate Cancer Cell-Matrix Interactions, Motility and Transendothelial Migration. PLoS ONE 2012, 7, e40594. [Google Scholar] [CrossRef] [PubMed]
- Niemantsverdriet, M.; Wagner, K.; Visser, M.; Backendorf, C. Cellular functions of 14-3-3ζ in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene 2007, 27, 1315–1319. [Google Scholar] [CrossRef]
- Liang, R.; Chen, X.-Q.; Bai, Q.-X.; Wang, Z.; Zhang, T.; Yang, L.; Dong, B.-X.; Gao, G.-X.; Gu, H.-T.; Zhu, H.-F. Increased 14-3-3ζ Expression in the Multidrug-Resistant Leukemia Cell Line HL-60/VCR as Compared to the Parental Line Mediates Cell Growth and Apoptosis in Part through Modification of Gene Expression. Acta Haematol. 2014, 132, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, J.; Du, Y.; Park, H.R.; Sun, S.-Y.; Bernal-Mizrachi, L.; Aitken, A.; Khuri, F.R.; Fu, H. Down-regulation of 14-3-3ζ suppresses anchorage-independent growth of lung cancer cells through anoikis activation. Proc. Natl. Acad. Sci. USA 2008, 105, 162–167. [Google Scholar] [CrossRef]
- Aseervatham, J. Cytoskeletal Remodeling in Cancer. Biology 2020, 9, 385. [Google Scholar] [CrossRef]
- Silva, L.E.; Souza, R.C.; Kitano, E.S.; Monteiro, L.F.; Iwai, L.K.; Forti, F.L. Proteomic and Interactome Approaches Reveal PAK4, PHB-2, and 14-3-3η as Targets of Overactivated Cdc42 in Cellular Responses to Genomic Instability. J. Proteome Res. 2019, 18, 3597–3614. [Google Scholar] [CrossRef]
- Kobayashi, H.; Ogura, Y.; Sawada, M.; Nakayama, R.; Takano, K.; Minato, Y.; Takemoto, Y.; Tashiro, E.; Watanabe, H.; Imoto, M. Involvement of 14-3-3 Proteins in the Second Epidermal Growth Factor-induced Wave of Rac1 Activation in the Process of Cell Migration. J. Biol. Chem. 2011, 286, 39259–39268. [Google Scholar] [CrossRef]
- Chen, M.; Liu, T.; Xu, L.; Gao, X.; Liu, X.; Wang, C.; He, Q.; Zhang, G.; Liu, L. Direct Interaction of 14-3-3ζ with Ezrin Promotes Cell Migration by Regulating the Formation of Membrane Ruffle. J. Mol. Biol. 2014, 426, 3118–3133. [Google Scholar] [CrossRef]
- Takala, H.; Nurminen, E.; Nurmi, S.M.; Aatonen, M.; Strandin, T.; Takatalo, M.; Kiema, T.; Gahmberg, C.G.; Ylänne, J.; Fagerholm, S.C.; et al. β2 integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood 2008, 112, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Deakin, N.O.; Bass, M.D.; Warwood, S.; Schoelermann, J.; Mostafavi-Pour, Z.; Knight, D.; Ballestrem, C.; Humphries, M.J. An integrin-α4–14-3-3ζ–paxillin ternary complex mediates localised Cdc42 activity and accelerates cell migration. J. Cell Sci. 2009, 122, 1654–1664. [Google Scholar] [CrossRef] [PubMed]
- Bonet, R.; Vakonakis, I.; Campbell, I.D. Characterization of 14-3-3-ζ Interactions with Integrin Tails. J. Mol. Biol. 2013, 425, 3060–3072. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, A.; Tanner, K.; Wang, D.; Geyer, F.C.; Reis-Filho, J.S.; Bissell, M.J. 14-3-3σ stabilizes a complex of soluble actin and intermediate filament to enable breast tumor invasion. Proc. Natl. Acad. Sci. USA 2013, 110, E3937–E3944. [Google Scholar] [CrossRef]
- Robens, J.M.; Yeow-Fong, L.; Ng, E.; Hall, C.; Manser, E. Regulation of IRSp53-Dependent Filopodial Dynamics by Antagonism between 14-3-3 Binding and SH3-Mediated Localization. Mol. Cell. Biol. 2010, 30, 829–844. [Google Scholar] [CrossRef]
- Kast, D.J.; Dominguez, R. IRSp53 coordinates AMPK and 14-3-3 signaling to regulate filopodia dynamics and directed cell migration. Mol. Biol. Cell 2019, 30, 1285–1297. [Google Scholar] [CrossRef]
- Nagata-Ohashi, K.; Ohta, Y.; Goto, K.; Chiba, S.; Mori, R.; Nishita, M.; Ohashi, K.; Kousaka, K.; Iwamatsu, A.; Niwa, R.; et al. A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J. Cell Biol. 2004, 165, 465–471. [Google Scholar] [CrossRef]
- Gohla, A.; Bokoch, G.M. 14-3-3 Regulates Actin Dynamics by Stabilizing Phosphorylated Cofilin. Curr. Biol. 2002, 12, 1704–1710. [Google Scholar] [CrossRef]
- Birkenfeld, J.; Betz, H.; Roth, D. Identification of cofilin and LIM-domain-containing protein kinase 1 as novel interaction partners of 14-3-3zeta. Biochem. J. 2003, 369, 45–54. [Google Scholar] [CrossRef]
- Ji, Z.-M.; Yang, L.-L.; Ni, J.; Xu, S.-P.; Yang, C.; Duan, P.; Lou, L.-P.; Ruan, Q.-R. Silencing Filamin A Inhibits the Invasion and Migration of Breast Cancer Cells by Up-regulating 14-3-3σ. Curr. Med. Sci. 2018, 38, 461–466. [Google Scholar] [CrossRef]
- Eiseler, T.; Hausser, A.; De Kimpe, L.; Van Lint, J.; Pfizenmaier, K. Protein Kinase D Controls Actin Polymerization and Cell Motility through Phosphorylation of Cortactin. J. Biol. Chem. 2010, 285, 18672–18683. [Google Scholar] [CrossRef] [PubMed]
- Abdrabou, A.; Brandwein, D.; Wang, Z. Differential Subcellular Distribution and Translocation of Seven 14-3-3 Isoforms in Response to EGF and During the Cell Cycle. Int. J. Mol. Sci. 2020, 21, 318. [Google Scholar] [CrossRef]
- Toshima, J.Y.; Toshima, J.; Watanabe, T.; Mizuno, K. Binding of 14-3-3β Regulates the Kinase Activity and Subcellular Localization of Testicular Protein Kinase 1. J. Biol. Chem. 2001, 276, 43471–43481. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Q.; Yu, A.C.H. The association of 14-3-3γ and actin plays a role in cell division and apoptosis in astrocytes. Biochem. Biophys. Res. Commun. 2002, 296, 657–663. [Google Scholar] [CrossRef]
- Moreno-Vicente, R.; Pavón, D.M.; Martín-Padura, I.; Català-Montoro, M.; Díez-Sánchez, A.; Quílez-Álvarez, A.; Lopez, J.A.; Sanchez-Alvarez, M.; Vázquez, J.; Strippoli, R.; et al. Caveolin-1 Modulates Mechanotransduction Responses to Substrate Stiffness through Actin-Dependent Control of YAP. Cell Rep. 2018, 25, 1622–1635.e6. [Google Scholar] [CrossRef]
- Freeman, A.K.; Morrison, D.K. 14-3-3 Proteins: Diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011, 22, 681–687. [Google Scholar] [CrossRef]
- Rushworth, L.K.; Hindley, A.D.; O’Neill, E.; Kolch, W. Regulation and Role of Raf-1/B-Raf Heterodimerization. Mol. Cell. Biol. 2006, 26, 2262–2272. [Google Scholar] [CrossRef]
- Aslan, J.E.; You, H.; Williamson, D.M.; Endig, J.; Youker, R.T.; Thomas, L.; Shu, H.; Du, Y.; Milewski, R.L.; Brush, M.H.; et al. Akt and 14-3-3 Control a PACS-2 Homeostatic Switch that Integrates Membrane Traffic with TRAIL-Induced Apoptosis. Mol. Cell 2009, 34, 497–509. [Google Scholar] [CrossRef]
- Kakinuma, N.; Roy, B.C.; Zhu, Y.; Wang, Y.; Kiyama, R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K–Akt signaling. J. Cell Biol. 2008, 181, 537–549. [Google Scholar] [CrossRef]
- Sandí, M.-J.; Marshall, C.B.; Balan, M.; Coyaud, É.; Zhou, M.; Monson, D.M.; Ishiyama, N.; Chandrakumar, A.A.; La Rose, J.; Couzens, A.L.; et al. MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity. Sci. Signal. 2017, 10, aan3286. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Arvanitis, D.A.; Eliopoulos, A.G.; Kranias, E.G.; Sanoudou, D. The Cardioprotective PKA-Mediated Hsp20 Phosphorylation Modulates Protein Associations Regulating Cytoskeletal Dynamics. Int. J. Mol. Sci. 2020, 21, 9572. [Google Scholar] [CrossRef] [PubMed]
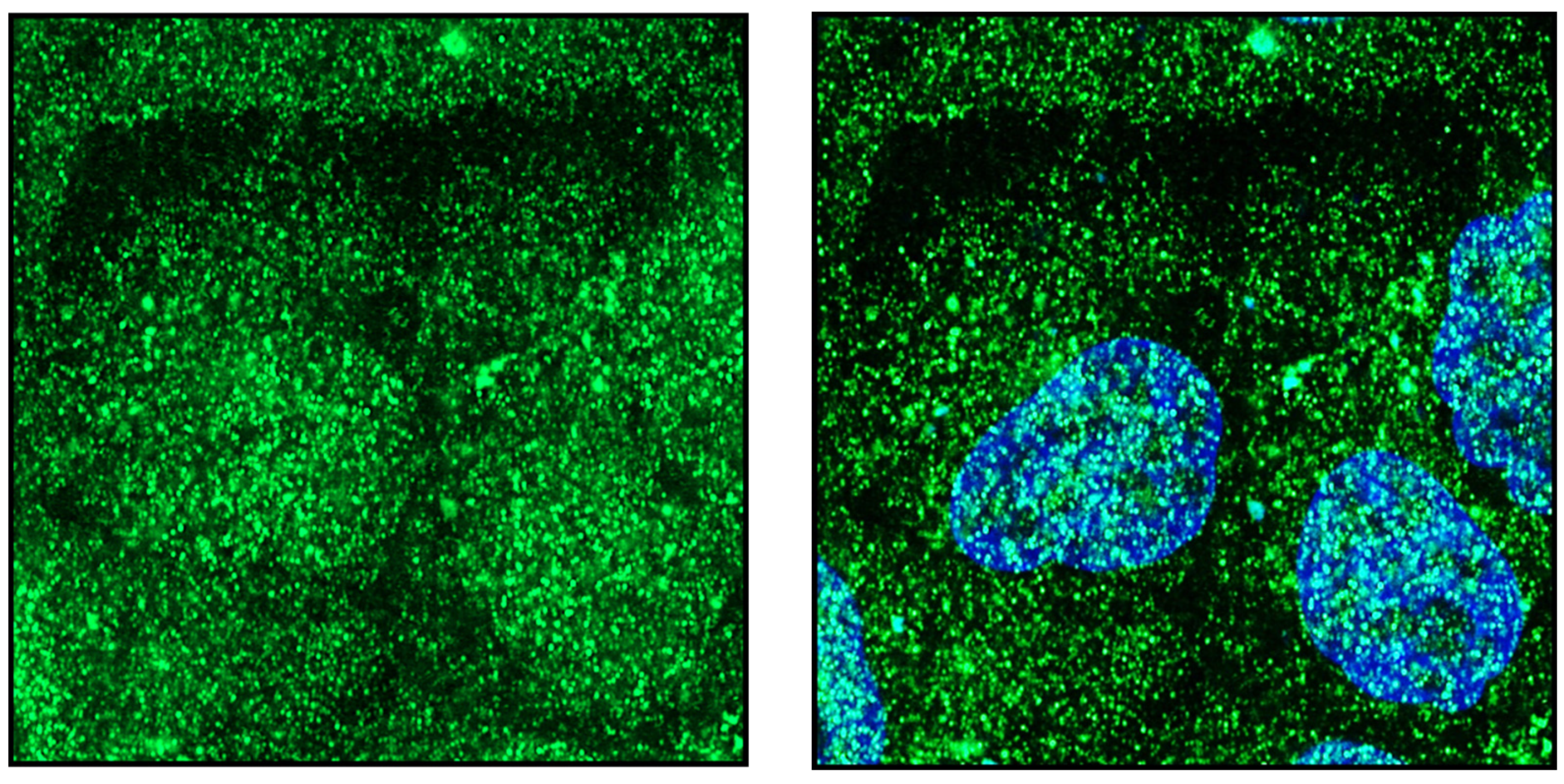
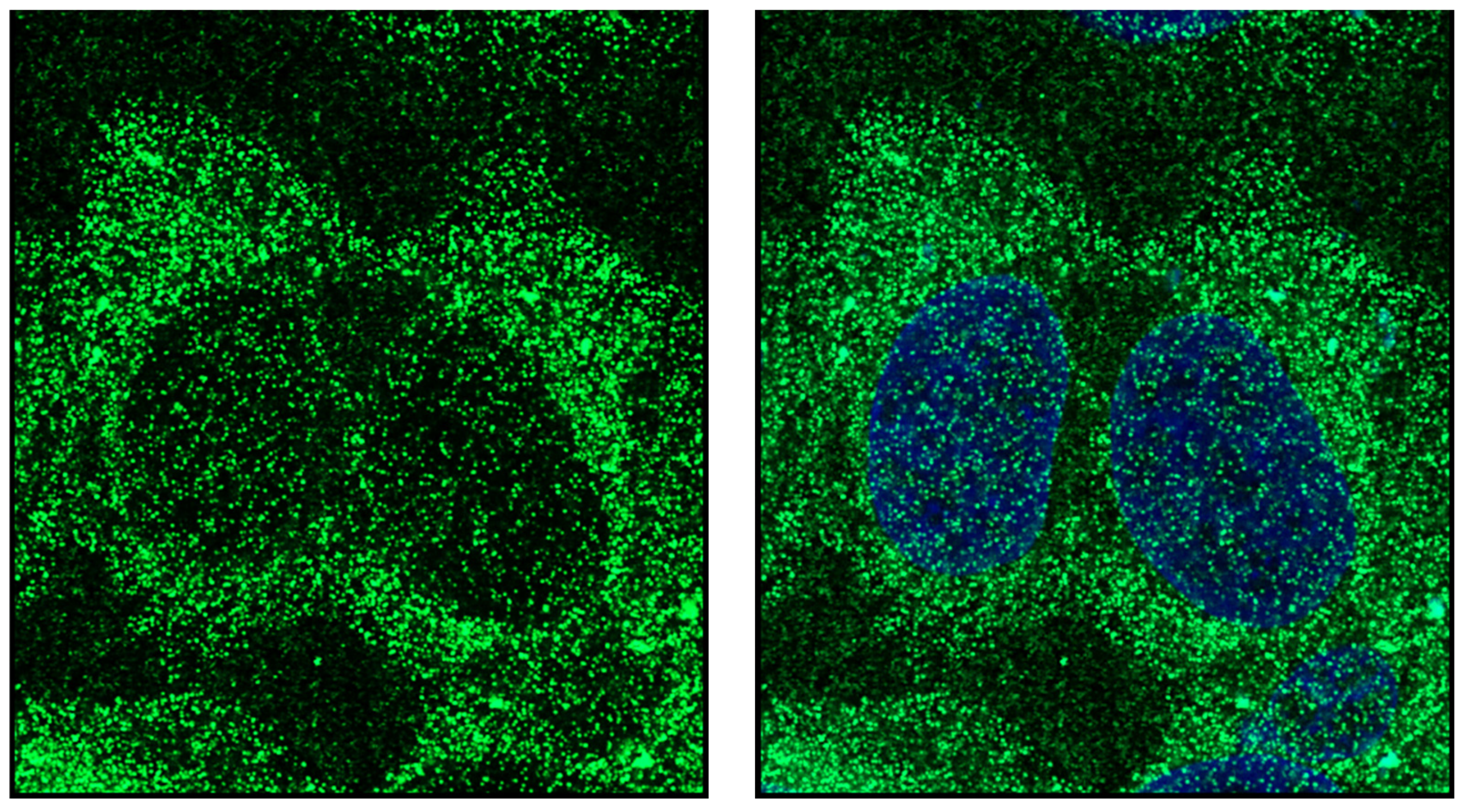
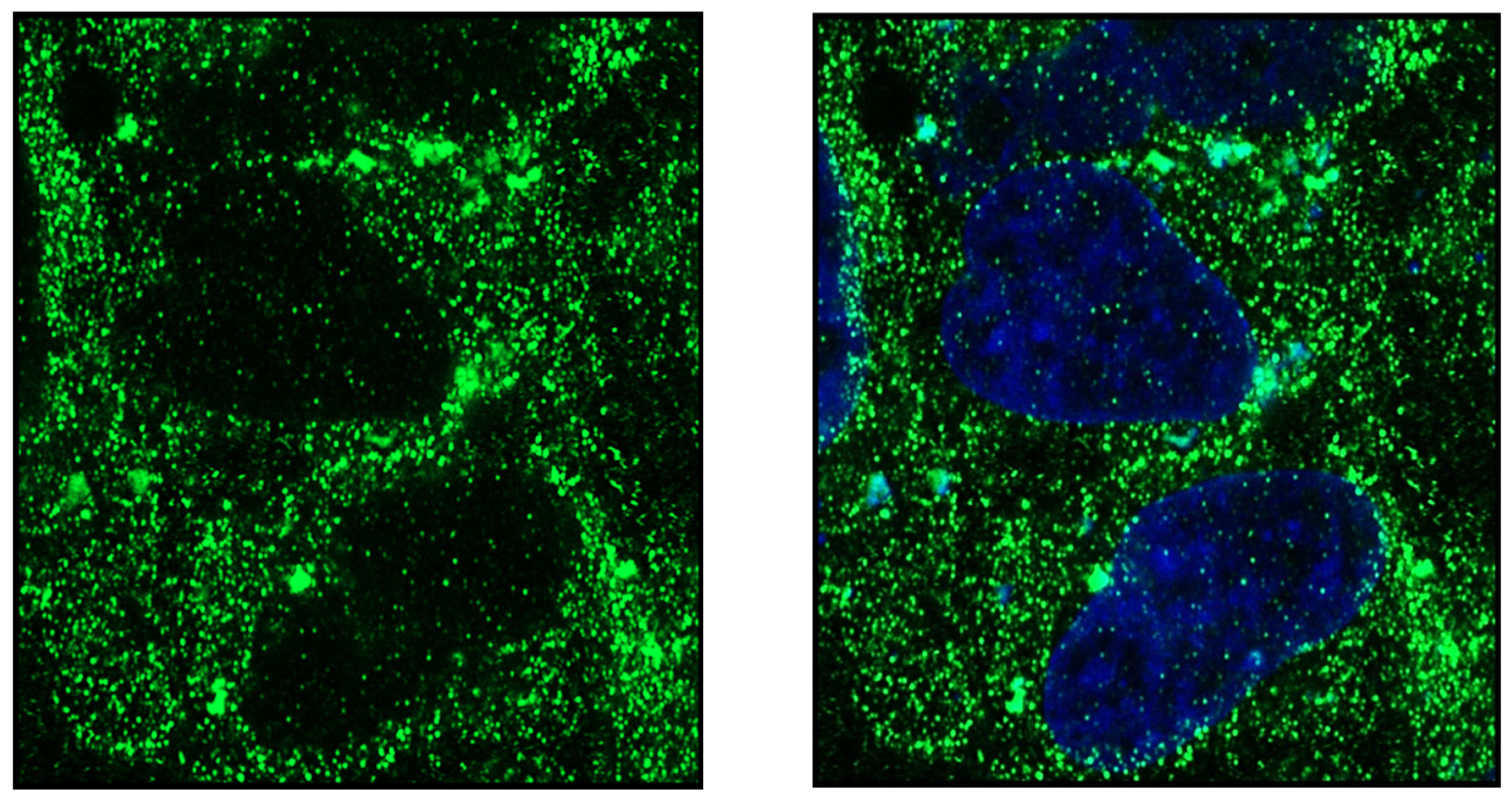
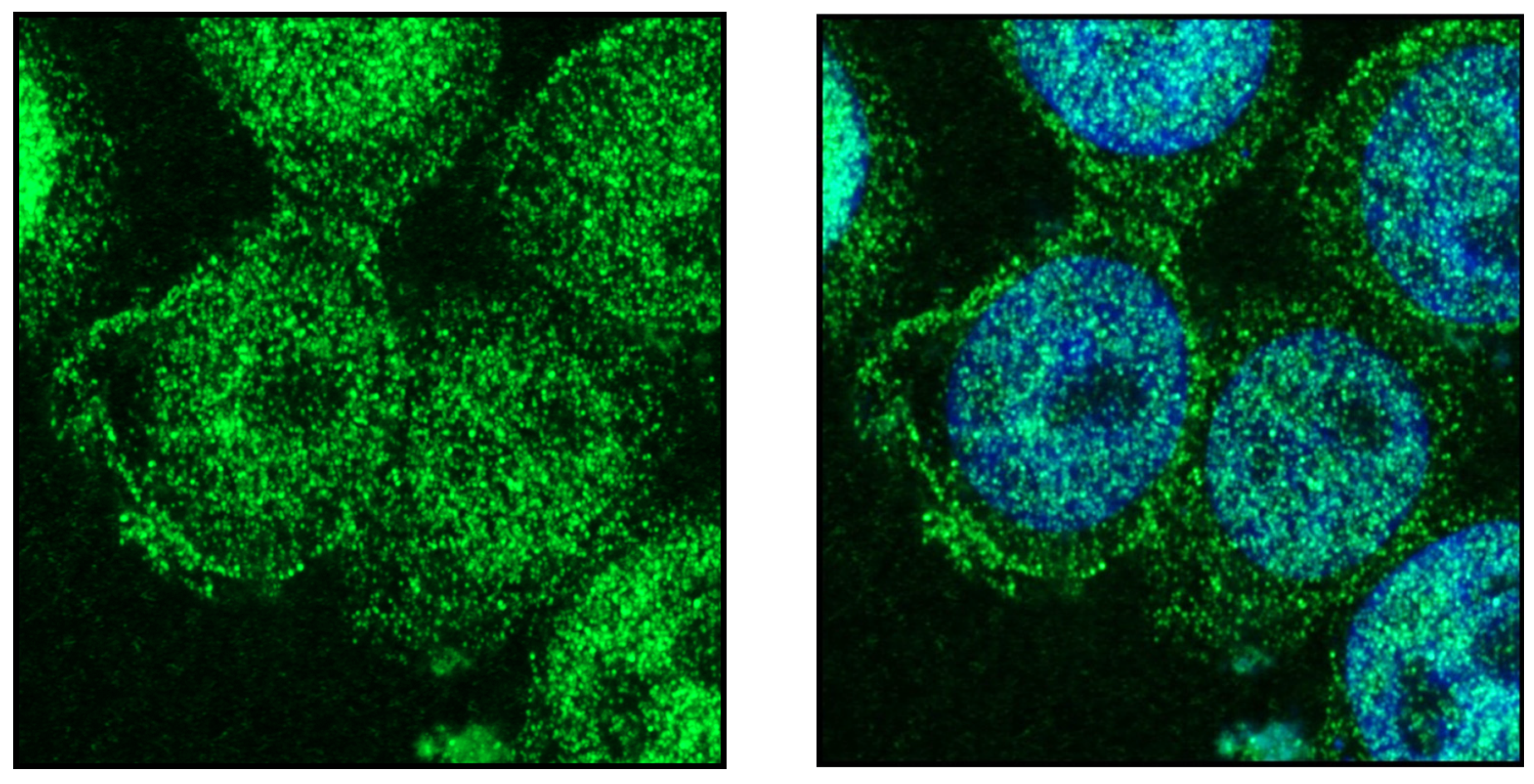
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aseervatham, J. Interactions between 14-3-3 Proteins and Actin Cytoskeleton and Its Regulation by microRNAs and Long Non-Coding RNAs in Cancer. Endocrines 2022, 3, 665-702. https://doi.org/10.3390/endocrines3040057
Aseervatham J. Interactions between 14-3-3 Proteins and Actin Cytoskeleton and Its Regulation by microRNAs and Long Non-Coding RNAs in Cancer. Endocrines. 2022; 3(4):665-702. https://doi.org/10.3390/endocrines3040057
Chicago/Turabian StyleAseervatham, Jaya. 2022. "Interactions between 14-3-3 Proteins and Actin Cytoskeleton and Its Regulation by microRNAs and Long Non-Coding RNAs in Cancer" Endocrines 3, no. 4: 665-702. https://doi.org/10.3390/endocrines3040057
APA StyleAseervatham, J. (2022). Interactions between 14-3-3 Proteins and Actin Cytoskeleton and Its Regulation by microRNAs and Long Non-Coding RNAs in Cancer. Endocrines, 3(4), 665-702. https://doi.org/10.3390/endocrines3040057




