Abstract
Prader–Willi Syndrome (PWS, OMIM #176270) is a rare complex genetic disorder due to the loss of expression of paternally derived genes in the PWS critical region on chromosome 15q11-q13. It affects multiple neuroendocrine systems and may present failure to thrive in infancy, but then, hyperphagia and morbid obesity starting in early childhood became the hallmark of this condition. Short stature, hypogonadism, sleep abnormalities, intellectual disability, and behavioral disturbances highlight the main features of this syndrome. There have been a significant number of advances in our understanding of the genetic mechanisms underlying the disease, especially discoveries of MAGEL2, NDN, MKRN3, and SNORD116 genes in the pathophysiology of PWS. However, early diagnosis and difficulty in treating some of the disease’s most disabling features remain challenging. As our understanding of PWS continues to grow, so does the availability of new therapies and management strategies available to clinicians and families.
1. Introduction
First described by Langdon Down in 1887 [1] and later by Prader, Labhart, and Willi in 1956 [2]. Prader–Willi Syndrome (PWS) is a rare, complex genetic disorder involving a maternally imprinted region on the proximal arm of chromosome 15 [3,4].
PWS affects multiple neuroendocrine systems with varying clinical features throughout affected individuals’ lives (Phases 0–4).
Phase 0, prenatal phase, is characterized with decreased fetal activity, breech presentation, polyhydramnios, and intrauterine growth restriction [5].
Phase 1 describes a period from birth to 25 months and is defined by initial hypotonia and poor feeding, which may require the use of a feeding tube or other methods of assisted feeding. Starting around 9 months, the infant begins to feed more appropriately and grows at a normal rate as measured by a growth curve.
Phase 2 occurs from approximately 25 months to 8 years, and the patient initially starts to cross percentiles of weight on a growth curve without an increased caloric intake. At around 4.5 years of age, the patient begins to display an increased appetite compared to their peers with continued crossing of weight percentiles if allowed to eat to satiation.
Phase 3 occurs after age 8 to adulthood, and it is in this time period that the patient develops an insatiable appetite and an obsessive fixation on food. Patients in this stage may exhibit food seeking behaviors in a manner similar to a patient experiencing substance addiction. Lying, stealing, placing oneself in dangerous situations to obtain food, and having emotional outbursts when food is denied have all been observed as a part of food seeking behavior in PWS. Drastic oversight of feeding behaviors, such as placing food under lock and key, may be needed to prevent hyperphagia-related injury or the exacerbation of obesity. Complications of morbid obesity and hyperphagia related injury are major concerns for patients in Phase 3.
Phase 4, which is only experienced by a small sub-set of PWS patients, shows a marked improvement of appetite control and no longer displays an intense preoccupation with food and shows the ability to feel satiated after eating [6].
Short stature, behavioral abnormalities, and sleep–wake cycle disruption result from multi-endocrine dysfunction that defines the syndrome. The incidence of the disease is estimated to be between 1/10,000 and 1/45,000 live births, and the average life expectancy is 29.5 years [3,7,8]. There is no difference in incidence based on sex, and differences in incidence based on ethnicity are too widely varied to draw a conclusion. Generally, the early cause of death is described as obesity-related, either with respect to medication complications due to morbid obesity or as accidents resulting from a pathological drive to eat. Currently, PWS is the most commonly known cause of syndromic obesity and the most common cause of obesity-related mortality [7].
The goal of this review is to illustrate the genetic basis of PWS, identify clinical manifestations of the disease and present the best practices for diagnoses and management.
2. Genetics
The PWS Region
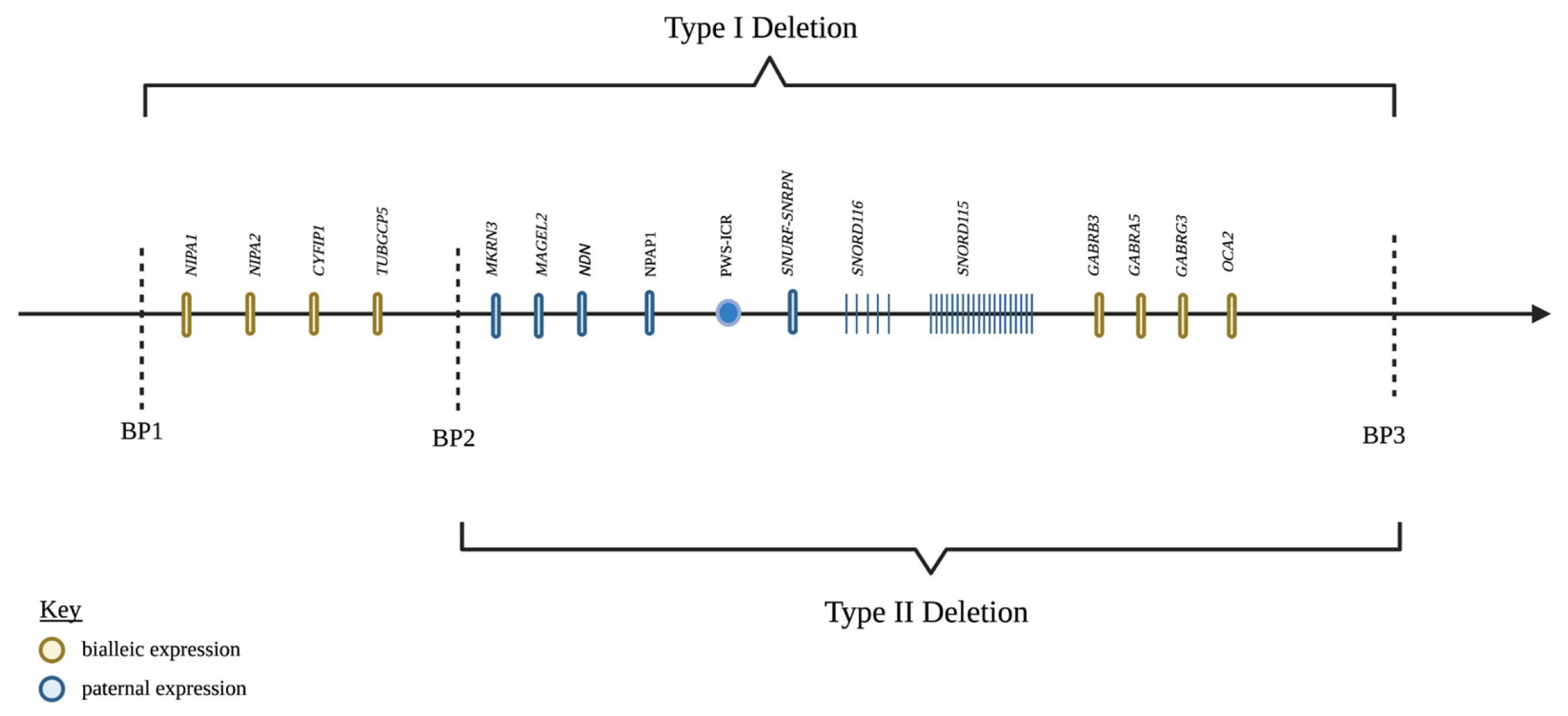
The region of the genome involved in PWS is 15q11-q13 (Figure 1). In a non-diseased state, this region is maternally imprinted, meaning the maternally inherited 15q11-q13 region is silenced and the paternally inherited region is expressed through epigenetic mechanisms [9,10,11]. The mechanism of silencing is carried out by various epigenetic modifications, such as the methylation of specific regions of DNA or histone tails and histone de-acetylation, which generally leads to a reduction in the expression of the genes in that region. Certain regions of imprinted gene clusters control the expression of the entire cluster, and these genes are referred to as imprinting control regions (ICR). Through phenomena are not completely understood, the ICR of PWS is differentially silenced in a parent-of-origin-dependent manner, meaning the paternal copy is expressed while the converse is true for the maternal copy. Thus, the silencing of PWS-ICR on the maternal chromosome leads to expression only on the paternal chromosome in non-diseased individuals. In simplest terms, PWS is caused by a loss of paternal expression of the genes in this region [12].
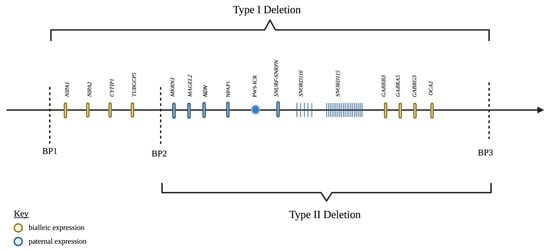
Figure 1.
Genes in the PWS critical region, chromosome 15q11.2-q13. Created with BioRender.com (accessed on 25 March 2022).
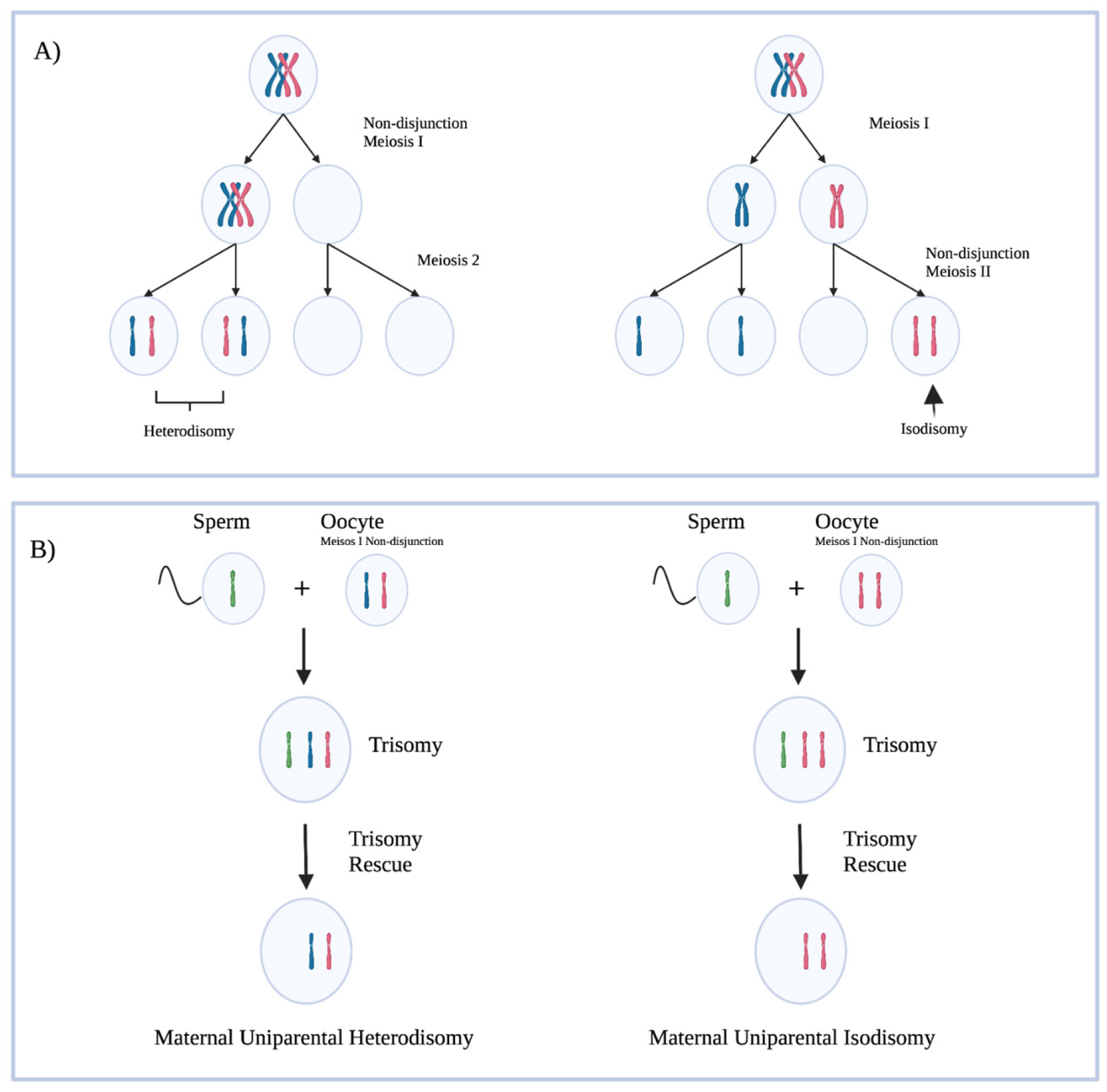
There are various ways in which the paternal PWS region may not be expressed. About 60–70% of PWS is caused by a deletion in the paternally inherited PWS locus, and 25–30% are due to maternal uniparental disomy (mUPD) (Figure 2) [13]. Maternal uniparental disomy has been shown to be more prevalent with increasing maternal age, which supports the hypothesis that non-disjunction events in meiosis lead to aberrant chromosome numbers, corrected by trisomy rescue. Approximately 4% of PWS is caused by imprinting defects caused by ICR microdeletions, epimutations not caused by deletion, and other non-characterized defects in imprinting [3].
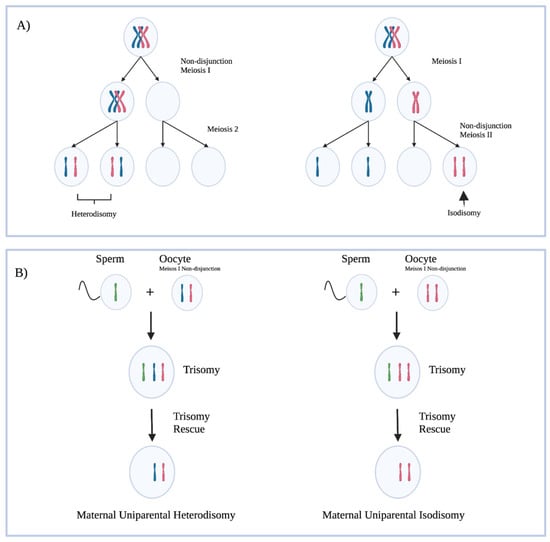
Figure 2.
(A) A representation of non-disjunction events in meiosis leading to abnormal chromosome copy numbers in gametes. Hetrodisomy refers to a gamete with each copy of a chromosome present. Isodisomy refers to a gamete with two copies of the same chromosome present. (B) Trisomy rescue leading to maternal uniparental disomy. Trisomy rescue in these cases does not always result in a maternal uniparental disomy in the cases outlined above; a maternal chromosome could have been lost. The outcomes of interest are displayed in the figure. Created with BioRender.com (accessed on 25 March 2022).
Amongst the paternal deletions, there are two major genetic subtypes with differing phenotypic expression classified as Type I and Type II deletions, which make up approximately 38.9% and 54.5% of the paternal deletion subtype, respectively [13]. To understand the different deletion subtypes, it is important to first understand the organization of the 15q11-q13 locus. The region of chromosome 15 is broken up into five break points (BP). The maternally imprinted genes of PWS are between BP2 and BP3. There are four non-imprinted genes (NIPA1, NIPA2, CYFIP1, and TUBGCP5) that reside between BP1 and BP [14,15,16]. Type I deletions consist of a loss of the genetic material between BP1 and BP3 (6.58 Mb), while Type II deletions involve the material between BP 2 and 3 (5.33 Mb) [17]. Butler et al. [18] were the first to describe a phenotypic difference between the two deletion types, with the Type I deletion exhibiting unique neuropsychiatric deficits not observed in Type II deletions. This difference suggested that the four genes in the BP1-BP2 region must be involved in neural functions in some way. Bittel et al. [19] showed that the amount of BP1-BP2 mRNA isolated from PWS patient-derived cell lines correlated strongly with the amount of phenotypic difference between the two deletion types. Further evidence was provided that all four genes were highly expressed in CNS. The identification of BP1-BP2 deletions independent of PWS would lend the most convincing support to Butler et al.’s findings of the phenotypic difference between type I and type II deletions [20,21]. The solitary BP1-BP2 deletion, or Burnside–Butler Syndrome, is characterized by intellectual disability and various neuropsychiatric disorders.
3. Updates to Genes of Interest in the PWS Region
Recently, advances have been made in the understanding of several genes implicated in the PWS phenotype. This review will cover recent information regarding MAGEL2, NDN, MKRN3, and SNORD116 (Table 1).

Table 1.
Summary of recent findings of genes implicated in PWS.
3.1. MKRN3
MKRN3, which encodes the E3 ubiquitin ligase makorin ring finger protein 3, is a maternally imprinted gene in the PWS region. The first evidence for a role of MKRN3 in the phenotype of PWS came from Abreu et al. [22], who identified a solitary deletion of this gene as a cause of central precocious puberty (CPP), thus linking it to the hypothalamic–pituitary–gonadal (HPG) axis. Currently, MKRN3 paternal inactivation is the most common genetic defect associated with CPP.
The molecular mechanisms underlying the initiation of puberty are not well understood, but it is generally accepted that the hormones that drive sexual development depend on a pulsatile secretion of GnRH from the hypothalamus. Early in life, there is a burst of GnRH followed by a period of quiescence until just before pubertal onset, when GnRH is again released in a pulsatile pattern. Abreu et al. [22] described that in mice and non-human primates the hypothalamic expression of MKRN3 decreases with the initiation of puberty. This, coupled with the link between MKRN3 and CPP, suggests that MKRN3 acts as a brake on the HPG axis. In a later study, Abreu et al. [23] discovered that MKRN3 is expressed within Kiss1 neurons, which are known to produce two stimulators of GnRH secretion: KISS1 and TAC3. MKRN3 was found to selectively repress the promotors of these two stimulatory genes, and as such act as a molecular brake on GnRH secretion. Patients with paternal mutations in the MKRN3 gene have not been shown to recapitulate the PWS phenotype on its own, but the newly elucidated role of the gene in the endocrine system suggests that it may provide contributions.
3.2. MAGEL2, NDN, and Circadian Rhythm
PWS patients are known to exhibit daytime sleepiness and night awakenings, which suggests there may be a role for PWS genes in the regulation of circadian rhythm. MAGEL2 and NDN have both recently been reported to have a role in circadian rhythmicity, but before we discuss their role, we will discuss the important factors driving normal rhythm [14,24]. The chief molecular components of circadian rhythm are the heterodimeric CLOCK:BMALI transcription factors, which control the expression of the Cry 1/2 and Period (Per) 1/2 genes. CRY and PER form a negative feedback loop w/ CLOCK:BMALI. Circadian rhythm involves the ubiquitination and degradation of CRY by SCF E3 ligases [31,32]. The direct role of CRY1 and CRY2 levels in rhythmicity is further borne out by the discovery that the inactivation of both abolishes rhythmicity, while their individual inactivation shortens or lengthens the circadian period, respectively.
MAGEL2 is a member of the melanoma antigen protein (MAGE) family, which interact with RING-zinc finger-type E3 ubiquitin ligases and deubiquitinases to modulate the ubiquitination of substrate proteins. In mice, MAGEL2 is expressed in a highly circadian rhythm, with a peak expression approximately 3 h before the peak of CRY1. MAGEL2 has also been found to be under the regulation of CLOCK, as the expression of MAGEL2 is altered in CLOCK mutant mice [24]. With this information in hand, it seems quite likely that there could be a role for MAGEL2 in modulating the ubiquitination of CRY1/2. Recently, Carias et al. [24] showed that MAGEL2 does in fact interact with SCF E3 ubiquitin ligases to mark CRY1 for degradation. This interaction is stabilized by USP7, which de-ubiquitinates CR1. Overall, it appears that MAGEL2, USP7, and SCF-E3 ligase complexes can all regulate CRY1 stability and that this process depends on the integrity of the MAGE homology domain. MAGEL2 interacts with USP7, but opposes its stabilizing effects, resulting in more ubiquitinated substrate and less CRY1 protein. These in vitro findings show that there is likely a role for MAGEL2 in circadian regulation, which may help explain the abnormal sleep–wake structure in PWS patients. However, in vivo studies of mice with a selective MAGEL2 deletion do not show a difference in CRY1 levels in the brain compared to WT mice.
NDN, a maternally imprinted intronless gene within the PWS region, which encodes the necdin protein, has been shown to be highly expressed in the suprachiasmatic nucleus (SCN), which is the pacemaker of the mammalian circadian clock. Mice deficient in necdin show abnormal sleep–wake behaviors and altered expressions of CLOCK-regulated genes. Lu et al. [14] showed that necdin interacts with BMALI, a core circadian protein, and a chaperone protein in complex to stabilize BMALI. The disruption of necdin expression destabilizes BMAL1 by promoting its proteolytic degradation through the ubiquitin–proteasomal system, which results in altered clock gene expressions and, thus, disrupted circadian rhythms. The role of NDN in circadian clock regulation provides evidence that, similarly to MAGEL2, it may play a role in the sleep–wake dysfunction observed in the PWS phenotype.
3.3. SNORD116
The smallest deletion syndrome that is known to fully recapitulate the PWS phenotype was first described by Sahoo et al. [27], who identified a patient with a deletion encompassing SNURF-SNRPN, part of SNORD115, and all of SNORD 116. Later, De Smith et al. [28] presented a patient with the PWS phenotype without any of the SNORD115 locus being implicated. This phenotype provided support for the hypothesis that SNORD116 is the minimal deletion required to cause PWS as, by this time, the literature had shown that both SNURF-SNRPN- [29,33] and SNORD115-specific perturbations were insufficient to replicate the phenotype.
SNORD116 is a sno-lncRNA without any currently described molecular function. Just as the human PWS phenotypes were observed in the deletion, congruent phenotypes were observed in mouse models of the same type. SNORD116 has been shown to be highly expressed in the mouse cortex, and in such a model, there is evidence that it may be an important regulator of epigenetic mechanisms [27,28,29,30]. It is crucial that future work aim to understand SNORD116 activity, as it currently stands as the PWS causative gene.
3.4. MAGEL2 and Schaaf-Yang Syndrome
Recently, a series of patients with nonsense mutations in MAGEL2 has been described in the literature as having a PWS-like syndrome (Schaaf–Yang Syndrome, SHFYNG) [25,26]. Similarly to PWS patients, these patients experience developmental delay, intellectual disability, neonatal hypotonia, failure to thrive in early childhood, and behavioral abnormalities. However, these patients do not experience the characteristic hyperphagia and morbid obesity that are observed in PWS. Furthermore, the paternal deletion of the MAGEL2 domain (that does not include SNORD116) results in a milder phenotype than the nonsense mutation of MAGEL2. Interestingly, the disruptions of USP7, which is known to interact with MAGEL2, produces a phenotype such as that observed in SHFYNG [24,25]. More work needs to be performed in order to identify the pathways where MAGEL2 is active, especially given that the modulation of this gene results in a PWS-like phenotype.
4. Clinical Presentation and Diagnostics
The diagnosis of PWS is complicated by its considerable range of phenotypic variance as well as the heterogeneity of genetic mechanism underlying the disease, thus centering it as the subject of considerable research interest. Initially identified solely by its clinical presentation, the genetic confirmation of PWS was first described by Ledbetter et al. [34] in 1981 as a deletion in the long arm of 15q11-q13 using high-resolution chromosomal analysis, and its diagnosis has since expanded to include a variety of genetic testing modalities [35].
There is considerable variability in age at diagnosis among the PWS patient population, spanning from birth to an upper bound of 20 to 48 years depending on the sample’s population, with a median of 0.3 years [36,37]. Testing technologies aim to definitively confirm a diagnosis of PWS in an accurate and economical manner earlier in a patient’s life (or prenatally), as earlier diagnosis is vital in assembling the significant health resources required for adequate management. Earlier diagnosis and treatment are associated with better outcomes in several measurements of PWS morbidity and is a central aim of PWS research [36,38,39].
The inherent variability in presentation and the wide range of non-specific clinical features associated with PWS generate a large differential diagnosis list when evaluating these patients including genital hypotonia, myotonic dystrophy, spinal muscular atrophy, fragile-X syndrome, and FG syndrome [40]. PWS that is identified later in life also necessitates the consideration of Klinefelter syndrome; Bardet–Biedl syndrome; Albright hereditary dystrophy; Börjeson–Forssman–Lehmann syndrome; maternal uniparental disomy chromosome 14 (Temple syndrome); copy number variants (CNVs) including del1p36, del2q37, and del6q16; Angelman syndrome; and Cohen syndrome, especially in cases in which the dominant feature is intellectual disability combined with non-specific obesity [40,41,42].
5. Clinical Criteria and the Diagnostic Pathway
Although the definitive diagnosis of PWS is only conducted by genetic analysis, its clinical manifestations are what lead to initial medical referral and consideration for testing. Several iterations of clinical diagnostic criteria exist for PWS, most of which currently build upon the 1993 consensus criteria [43], which assigns point values to major and minor criteria to arrive at a diagnosis. Major criteria include infantile hypotonia that gradually improves with age, feeding problems and failure to thrive, which transitions to rapid weight gain first appearing between 12 months and 6 years of age, hypogonadism, and characteristic facial dysmorphias such as narrow bifrontal diameter, almond-shaped eyes, and a thin upper lip. Minor criteria include sleep disturbances, short stature, characteristic behavioral problems like temper tantrums and obsessive-compulsive behavior, hypopigmentation, small hands, and eye abnormalities such as esotropia and myopia. The 1993 criteria were initially diagnostic as they were implemented before genetic testing was widely available, whereas they are now only used to determine the appropriateness of further testing in developed nations. Clinical features are not enough to accurately diagnose PWS on their own, as no one symptom or a constellation of symptoms is specific to an individual mechanistic subtype [44]. An overreliance on clinical criteria for the diagnosis and management of PWS can be problematic, as a retrospective study of 111 suspected pediatric PWS cases in Cuba showed, in which only 50% of the clinically suspected cases were confirmed genetically via FISH [45].
With the advent of widespread available diagnostic genetic testing, the 1993 criteria were modified to stratify by age to reflect the specific disease manifestations most prominent at each stage of development, increasing access to definitive testing by correcting for the non-specificity of symptoms and their tendency to evolve subtly over time specifically features of neuromuscular deficiency as the salient manifestation in early childhood and hyperphagia and cognitive impairment becoming more prominent later in development [46]. In a sample of 68 PWS patients, 17% of the confirmed PWS patients did not meet the 1993 criteria’s threshold for diagnosis, and several of the minor criteria (e.g., decreased fetal activity, behavioral problems, and sleep apnea) were more diagnostically sensitive than the major criteria (e.g., characteristic facial features) [46]. Recently described clinical phenomena in the broader constellation of PWS symptoms include partial ankyloglossia, median grooved tongue [47], and rare pediatric heart tumors such as cardiac rhabdomyoma [48]. A reasonable diagnostic heuristic for the community physician is that any infant with hypotonia is likely a good candidate for PWS genetic screening [46].
Once adequate clinical suspicion for PWS exists, patients are referred for the preferred screening test, methylation analysis, followed by confirmatory testing to detect the subtype that has classically been karyotyping with FISH to reveal deletions and rearrangements on chromosome 15 [40,49,50]. As per the 2019 update of the EMQN/ACGS best practice guidelines for the molecular analysis of PWS [51], initial methylation screening should be performed either through methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) across the 15q11-q13 region or methylation-specific polymerase chain reaction (MS-PCR) at the SNRPN locus. Methylation testing is the only diagnostic method able to identify PWS caused by the three primary pathogenic mechanisms (paternal deletion, maternal UPD, and imprinting defects) [52], identifying >99% of PWS and 80% of Angelman Syndrome. MS-MLPA will identify the deletion status but will not differentiate between UPD and ID, while MS-PCR fails to differentiate between mechanistic subtypes altogether [52]. Likewise, FISH may have limited utility in detecting UPD and lookalike conditions such as UPD of chromosome 14 [45] as well as other unique SNURF-SNURPN missense variations associated with PWS-like syndromes [53]. Alternative confirmation methods include short tandem repeat analysis (STR), comparative genetic hybridization (CGH), and chromosomal microarray analysis (CMA) [54]. If screening and confirmatory testing are negative but suspicion for PWS remains high based on clinical presentation, an imprinting defect through epigenetic mutation may be suspected [50]. The above screening and confirmation diagnostic paradigm forgo unnecessary and invasive muscle and nerve biopsies.
6. Emerging Technologies in PWS Diagnosis
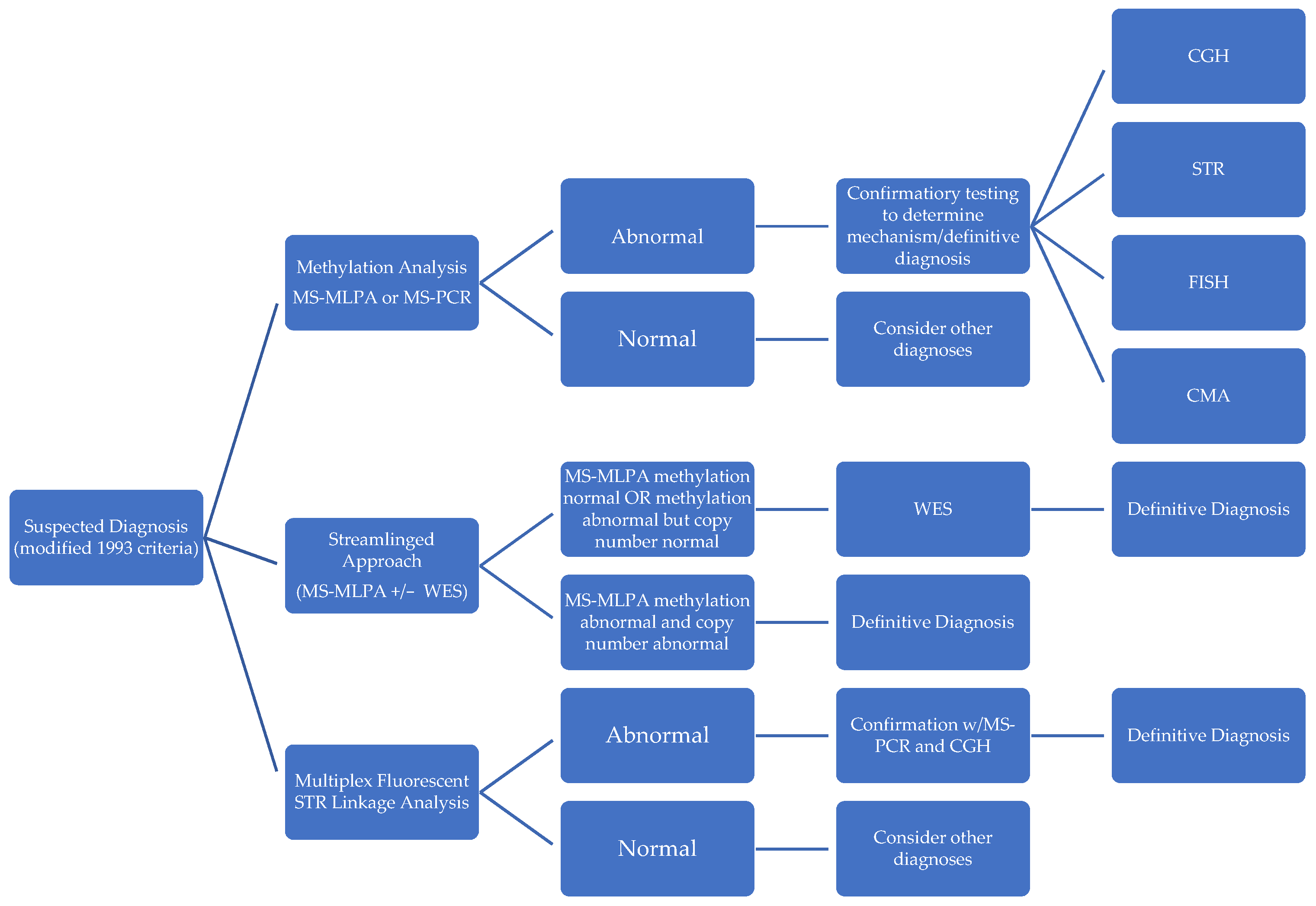
To address challenges in the affordability, speed, and accuracy of subtype identifications in PWS diagnosis, several novel methodologies have been described in recent literature (Figure 3).
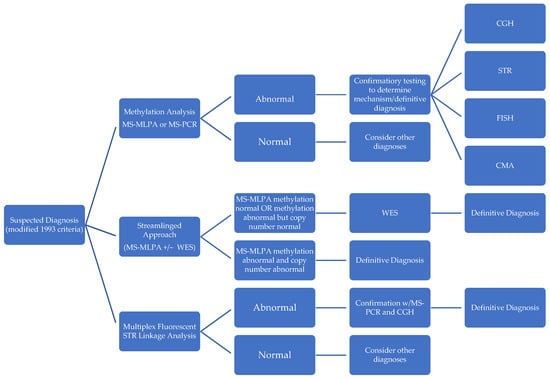
Figure 3.
Comparison of diagnostic approaches in PWS.
In contrast to the standardized stepwise approach of first performing a screening methylation test and then following up with further confirmatory tests, Strom et al. [41] constructed a “streamlined molecular approach” that efficiently determines subtype and negates the requirement for further diagnostic testing. They accomplish this by using methylation-sensitive MLPA combined with whole exome sequencing (WES) with sequence variant analysis of SNRPN and UBE3a, copy number analysis, and absence of heterozygosity analysis on chromosome 15 for diagnosis and mechanism in a single report. This approach entails deferring to WES when MS-MLPA reveals no methylation (to rule out PWS-like disorders), as well as when MS-MLPA methylation detection is positive, but copy number detection yields a normal copy number (to identify microdeletions, atypical deletions, disomy status, and UPD subclass). MS-MLPA is a variant of multiplex PCR that uses a single primer for multiple targets enabling the detection of copy number changes as well as methylation status and was shown to be highly accurate in detecting PWS with a 100% concordance with MS-PCR-based diagnosis in a study of 98 patients [55]. However, unlike MS-PCR, it has the distinct advantage of being able to differentiate between deletion and non-deletion subtypes of PWS, but it is unable to distinguish UPD from imprinting defects [56]. In combination with whole exome sequencing, this streamlined approach identifies mechanisms much more precisely than using MS-PCR alone, detecting mosaicism, segmental versus total isodisomy 15, additional X-linked disorders in females from skewed X-inactivation, Rett syndrome, as well as copy number variants and point-mutations in other causative genes. It could also be further modified to include an analysis of chromosome 14 to rule out alternative diagnostic considerations such as Bardet–Biedl syndrome and Cohen syndrome, which would be informative in 97% of patients [41]. Additionally, unlike other methods, it requires no parental DNA to identify imprinting center microdeletions. Techniques that employ whole exome sequencing have the highest diagnostic yield behind invasive muscle biopsy [57] and they could be vital in genetic counseling (e.g., imprinting defects raise risk of having another child with PWS to 50%) [37] and surveillance for associated comorbidities. Potential barriers to the widespread adoption of whole exome sequencing combined with MS-MLPA include its cost, technical complexity, and non-uniform coverage by health insurance systems worldwide.
Zhang et al. [54] likewise described another novel diagnostic methodology: multiplex fluorescent STR linkage analysis, which identifies mechanism and subtype of PWS. Using multiplex fluorescent labeled PCR in STR linkage analyses, the results are subsequently validated by MS-PCR and array CGH. This technique, such as MS-MLPA plus whole exome sequencing, provides mechanistic detail but with a distinct advantage in turnaround time when compared to alternative testing methods: 5 h vs. 2 days for MS-PCR or MS-MLPA, 3 days for CGH, and 5 days for STR direct sequence analysis. Other common tests do not provide complete information: CMA cannot detect heterodisomy, MS-PCR cannot differentiate subtypes, MS-MLPA cannot differentiate UPD vs. imprinting defects, and STR direct sequencing cannot differentiate between types of deletion and uniparental isodisomy [58]. Zhang et al.’s study of 26 patients in China showed multiplex fluorescent STR linkage analysis to be accurate and economical when compared with alternative methodologies.
7. Prenatal Diagnosis
The reliable prenatal diagnosis of PWS has remained an elusive goal that is the focus of current research efforts. Prenatal testing is usually limited to families in which there is a known history of PWS or otherwise suspicion for an imprinting center deletion or chromosomal rearrangement. Recurrence varies widely between mechanisms of disease. If the father has an imprinting center deletion or if there is an unbalanced chromosomal rearrangement, recurrence can approach 50%. However, the risk of recurrence in mUPD or de novo deletions is very low [59]. There are a small number of reports in the literature of successful incidental prenatal PWS diagnoses, typically the product of suspicious findings on ultrasound that lead to further genetic testing [52], as microdeletion and methylation detection are not typically performed in the normal screening for maternal genetic syndromes in amniocentesis or chorionic villus sampling (CVS) [46].
Maternal and fetal characteristics indicative of PWS include abnormal fetal growth, decreased fetal movements, anomalous extremity positions, polyhydramnios, cryptorchidism, fetal goiter, cardiac rhabdomyoma, breech presentation, hospitalization during pregnancy, advanced maternal age, decreased birth weight, preterm birth, and nearly universally normal ultrasound findings before the 24th week of gestation, although they are often highly variable and can be completely undetectable prenatally [35,38,48,60]. Many of the main prenatal features of PWS may be a product of neuromotor developmental dysfunction and the resulting decreased swallowing of amniotic fluid [48]. However, none of these clinical features are specific to PWS and are not adequate on their own for diagnosis. Further complicating early diagnosis via ultrasound may be a bias among healthcare professionals toward trying to obtain normal sonography results, even in the face of maternal complaint, which may further delay appropriate testing [61]. A retrospective analysis of 101 pregnancies referred to a PWS treatment center postnatally revealed that only one of the women had MS-PCR performed before birth, even though many had distinct prenatal findings [38].
Even when an abnormality is suspected, testing may not positively identify PWS (Table 2). Chromosomal microsomal analysis, a common first-tier test for neurodevelopmental disorders in utero may fail to detect complete hetero-UPD, which may have less severe (later hyperphagia, higher birthweights, and less hypotonia) symptoms that are easier to miss upon diagnosis [52,56,62]. Dong et al. [52] described the case of an infant boy who had many of the typical PWS features upon ultrasound in utero but passed CMA testing and chromosome analysis. He was then formally diagnosed using CMA and MS-MLPA after birth, eventually succumbing to his illness at the age of 4 months.

Table 2.
Comparison of prenatal diagnostic strategies for PWS.
Prenatal clinical indications of PWS, especially the highly suspicious combination of decreased fetal growth with normal doppler studies and polyhydramnios (ruling out the most common cause of decreased fetal growth: placental insufficiency) [63], should be referred for further testing. Traditionally, amniocentesis or chorionic villus sampling may be performed, along with methylation screening and further confirmatory studies for mechanism and subtype [35].
Considerable interest surrounds the possible application of non-invasive prenatal screening (NIPS) toward the prenatal detection of PWS. First used for detecting aneuploidies, NIPS uses fetal DNA in maternal plasma as a source for testing through either parallel DNA sequencing or SNP-based studies that analyze maternal and fetal gene distributions [35]. This can be performed as early as 9–10 weeks’ gestation with results being ready in 7–10 days [35]. Shubina et al. [64] described the first case of PWS diagnosed with NIPS using high-throughput shallow whole genome sequencing confirmed via amniocentesis with FISH analysis and an SNP-based chromosomal microarray [64]. PWS was diagnosed successfully, and pregnancy was terminated at 21 weeks. NIPS represents a viable PWS screening method that is considerably less invasive than the alternatives. However, its diagnostic utility is not as precise as other methods when SNP microarrays are used as a confirmation method, as it likely does not detect UPD. Still, 6% of women with a suspected positive prenatal diagnosis of PWS on NIPS choose to terminate without seeking further confirmatory tests [35]. Additionally, when NIPS is used in microdeletion detection, it suffers from a high rate of false positives through its high sensitivity in identifying variants of unknown significance, which has a unique potential to cause harm when employed in the sensitive prenatal period [37]. Mixed availability, varying policies on insurance reimbursement for the test, and ethical considerations are current barriers to its widespread adoption.
Patients undergoing NIPS screening could likely benefit through consultation with a medical geneticist, as the successful prenatal diagnosis of PWS not only makes new options available for earlier intervention but also provides pregnancy management. Approximately 50% of parents say they would support the termination of a pregnancy that was positive for PWS, even in the third trimester [65].
8. Management
The clinical management of PWS spans many therapeutic domains including nutritional, developmental, educational, hormonal, and behavioral support, with each stage of development requiring unique management strategies [40]. Features of the disease that contribute to significant morbidity and mortality likely stem from hypothalamic dysfunction [50], which include growth hormone deficiency and its numerous sequelae, hypothyroidism, obesity, diabetes mellitus, hyperlipidemia, obstructive sleep apnea, central adrenal insufficiency, hypogonadism, developmental disability, mood disorders, and nervous and musculoskeletal abnormalities, for which its management necessitates the use of a multidisciplinary care model [47,53,66,67].
Despite significant advances in our understanding of PWS, the disease remains a life-limiting condition with a mortality rate approximately 3 times of that of the general population [68]. One of the largest studies of PWS mortality [69] identified the median age at death as 30 years, ranging from 1 month to 58 years. The leading causes of death were identified as the result of the frequent morbid obesity observed in the PWS population, including restrictive (in adults) and infectious (in children) respiratory pathologies, as well as sudden death, cardiac failure, and pulmonary embolism.
8.1. Growth Hormone Therapy
Approved by the FDA in 2000 for PWS treatment in children, growth hormone (GH) replacement therapy is one of the primary treatment tools for PWS, possessing robust evidence for efficacy and safety [70]. Between half and ¾ of all children with PWS have a measurable GH deficiency, and nearly all have a deficiency in IGF-1 [67]. Treatment is started empirically as soon as PWS is suspected, at as early as 3 months whenever possible [67,71]. GH stimulation testing prior to initiation of therapy is unnecessary and should not be performed. Stimulation testing measures pituitary secretion which can be falsely elevated in PWS patients due to its hypothalamic involvement. Additionally testing may be difficult to perform in insulin-resistant patients who cannot achieve hypoglycemia [70].
The dosage of recombinant human growth hormone (rhGH) typically begins at 0.5 mg/m2/d titrating up to 1.0 mg/m2, with monthly IGF monitoring initially and extending to once every 6 months for longer-term surveillance. Polysomnography screening for obstructive sleep apnea should be completed before treatment initiation, 3–6 months after the start of treatment, and then annually [67]. Growth hormone improves muscle strength, linear growth, psychomotor development, metabolic state, cognition, and health-related quality of life [72,73]. GH is not recommended for those with uncontrolled diabetes, active malignancy, and acute critical illness.
Many of the commonly reported benefits of GH therapy in the literature were confirmed by Angulo et al. [74] in the largest-to-date sample of pediatric patients on rhGH (~40,000) over a 10-year period, of which 129 pediatric PWS patients met inclusion criteria. Longer durations of treatment and younger age at initiation of treatment were both associated with significant improvements in height standard deviation scores. The data also suggested that GH therapy has opposite effects on BMI during the different nutritional phases of PWS, increasing BMI during earlier phases of poor feeding and lowering BMI when patients enter later hyperphagic phases. Therapy was found to be well-tolerated with five serious reactions observed. Two deaths were reported which were likely not directly attributable to rhGH.
GH therapy for children found near-universal acceptance among health systems worldwide; a much smaller number of countries, including the USA, currently recognize the benefits of continuing therapy through adulthood [70]. In adults, GH therapy can improve body composition (without significantly changing BMI), fatigue, cognitive function (with regression upon discontinuation) [75], motor skills, muscle strength, cardiac risk profile, and general markers of quality of life. However, GH therapy does not improve bone mineral density [76] or thyroid function. GH appears to modulate T4, T3, and rT3 in non-PWS patients with GH deficiency but does not affect TSH levels upon TRH testing in PWS patients. There is no consensus on how to evaluate or treat hypothyroidism in the context of PWS [77]. Potential burdens and risks of GH therapy in children and adults include sleep apnea, increased blood sugar, and extra burden and reliance on caregivers as most PWS patients are not able to manage a consistent schedule of rhGH injections on their own.
8.2. Hyperphagia and Obesity
Hyperphagia and obesity, the uniquely debilitating calling-cards of PWS, affect 40% of children and adolescents with the disease and up to 98% of adults [78]. They present as uncontrollable urges to eat massive quantities of food, even those that are spoiled or frozen [79]. Other notable behaviors include the stealing of food, extreme frustration over food restrictions, and hoarding of food, all which become prominent around school age (median 8 years) after a primary phase of poor feeding and failure to thrive [80,81,82]. Ultimately, hyperphagia underlies most of the main mechanisms of morbidity and mortality in PWS and can even lead to direct gastric necrosis by its own right [80].
First-line therapies for hyperphagia and obesity management in PWS include aggressive behavioral and nutritional support through the strict limitation of access to food, close monitoring of caloric intake and types of foods consumed, and regular exercise [40]. Calories must be restricted to counteract the markedly diminished metabolic energy expenditure observed among PWS patients, even when compared to obese non-PWS individuals. Height ranging 8–11 kcal/cm comprises an ideal range for caloric intake to maintain a healthy weight. To lessen anxiety and fixation on food at mealtime, portion control should make use of the patient’s own palms or hands for measurement [80]. Counteracting hyperphagia implicitly places limitations on an individual’s autonomy and independence. The development of effective pharmacological therapies for the control of problem eating behaviors could theoretically enable individuals with PWS to live and function independently.
Topiramate is an antiepileptic drug and mood stabilizer that has seen recent experimental use as an appetite suppressant for PWS. It is known to have weight loss and appetite suppressing activity as a side effect and has been investigated as a potential treatment for disordered eating behaviors, such as bulimia nervosa. Previous small studies have offered conflicting evidence for its efficacy in treating PWS. The TOPRADER trial [82], a multi-center, double-blind, 8-week long RCT performed in France, studied the efficacy of topiramate in controlling eating and other behavioral pathologies in PWS. When combined with behavioral and educational resource support, Topiramate was found to have a significant effect on modulating hyperphagia but not on other behavioral symptoms such as skin picking and irritability. Topiramate was generally well tolerated with lethargy, psychomotor slowdown, and hepatic function decrease as adverse effects.
Caralluma Fimbriata is another potential therapy for hyperphagia in PWS. Caralluma is a wild shrub used in Indian folk medicine as an appetite suppressant. In PWS animal models, it has shown to enhance 5-HT2c receptor activity, the same receptor to which hyperphagia-associated SNORD116 and SNORD115 deletions are thought to interfere with in the hypothalamus [83]. A case report [84] of Caralluma therapy over 12 years in a 14-year-old female PWS patient was notable for a dramatic reduction in hyperphagia that permitted her to safely have freedom around food, which reverted to baseline upon the cessation of therapy. Weight loss was also observed, but similarly to Topiramate, her other PWS behavioral symptoms (including skin picking) were not attenuated. No major side effects were experienced. Interestingly, the patient drank plenty of water while receiving therapy in spite of individuals with PWS being relatively disinterested in water, often having to be coerced to drink by caregivers [80].
Glucagon-like peptide (GLP-1) agonists, originally used in controlling Type II diabetes mellitus, have found use as a potential therapy for PWS-related obesity. Kim et al. [80] describe the use of liraglutide in a morbidly obese 18-year-old female PWS patient hospitalized with restrictive respiratory failure, who after 2 years of therapy progressed from a BMI of 71 kg/m2 to 37 kg/m2 and was sated on an appropriate diet of 1000 kcal/day. A small number of case reports suggest promising results with GLP-1 agonists and warrant further studies into their use in PWS [85,86].
Other drugs currently being investigated for use in PWS hyperphagia and obesity control include naltrexone/bupropion (hypothalamic POMC activator), orlistat (pancreatic lipase inhibitor), sibutramine (nonspecific inhibitor of serotonin and norepinephrine reuptake), and rimonabant (endocannabinoid CB1 receptor antagonist) [81].
8.3. Behavioral Health
PWS presents with a constellation of behavioral disorders such as cognitive rigidity, anxiety, mood disorders, self-injurious behavior—especially in individuals with a deletion subtype—and affective psychosis and autism spectrum symptoms in those with UPD [40,87]. Behavioral disorders in PWS are frequently the target of psychopharmaceuticals, typically selective serotonin reuptake inhibitors (SSRIs) (50% of children and 70% of adolescents and adults) and atypical neuroleptics (34%). Polypharmacy is a common feature of behavioral management with significant adverse effect burdens, such as worsening obesity [80] and increasing risk of rhythm disorders that could lead to sudden cardiac death [69].
Forster et al. [87] used the largest sample of PWS patients to date to undergo pharmacogenetic testing in order to identify significant differences in metabolizing status among CYP450 enzymes that are the frequent targets of popular psychopharmaceuticals used in treatment. There are significant differences in drug response, adverse effects, medication interactions, and the likelihood of rehospitalization between ultra-rapid metabolizers, extensive metabolizers, intermediate metabolizers, and poor metabolizers. For example, with regards to CYP2D6, 17.1% of PWS patients were poor metabolizers compared to 2.2% in the general population, and 48.6% had extensive metabolizing compared to 63.6% in the general population. Ultrarapid metabolizers are more likely to have quicker and more positives responses to antidepressant therapy [88]. For any medication that is metabolized via CYP2D6, PWS patients might experience better outcomes employing a smaller-than-normal dose. Similar differences exist for CYPs 2C19, 2C9, 3A4, and 1A2. This could change the dosage calculus for drugs such as sertraline, modafinil, and aripiprazole and underscores the potential utility of pharmacogenetic testing in PWS management to reach better outcomes and fewer adverse effects when using psychopharmaceuticals.
Pitolisant is a strong histamine H3 receptor-mixed agonist/antagonist that is not FDA approved but has approval in Europe for use in narcolepsy. Pitolisant has found recent experimental use in treating PWS. Pennington et al. [89] described a 15-year-old female PWS patient with obsessive–compulsive disorder (OCD), daytime sleepiness, autism spectrum symptoms, and mild intellectual disability on stimulants, SSRIs, and atypical antipsychotics. Shortly after therapy began, fewer behavioral outbursts, improved muscle tone, and diminished hyperflexibility were observed, although she maintained her rigid thought processes. Twelve months post-initiation, depression, irritability, lethargy, and hyperactivity continued to improve, although separation anxiety and generalized anxiety worsened. Notably, she was able to taper and cease SSRI and buspirone therapy. Side effects comrpised a 2% increase in BMI, sweating, salivation, and drooling.
PWS patients are likely deficient in oxytocin via MAGEL2 [90] deficiency, which may be involved in some of the behavioral features of PWS. Oxytocin is important for social skills, food intake, and body weight. Damen et al. [91] performed a double-blinded placebo-controlled clinical trial investigating three months of intranasal oxytocin therapy among 26 children with PWS. Interestingly, positive effects were observed in males with PWS, such as a decrease in orexigenic drive, but not in females. Furthermore, patients with deletion mutations had a better response than patients with maternal UPD. More than half of the participants’ parents wanted to continue intranasal oxytocin off-label after the conclusion of the trial because of the positive effects on social behavior that were observed at home. No serious adverse events were observed during the trial period.
9. Future Developments
As with other genetic diseases for which the available treatments are primarily limited to symptom relief, genetic and epigenetic therapies provide future hope for a potential cure, particularly in PWS where every affected patient already has at least one functional copy of the silenced genes associated with its pathogenesis [92]. As of the time of writing this paper, genetic and epigenetic therapies for PWS have yet to advance to the clinical trial stage.
In general, gene therapy attempts to treat disease by replacing faulty genes, introducing new genes, or modifying faulty genes through the direct alteration of a living patient’s genetic material. A number of genome editing tools including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the RNA-guided CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein 9) nuclease system have been developed in recent years [93]. The first two tools bind endonuclease catalytic domains to modular DNA-binding proteins to induce targeted DNA double-stranded breaks (DSBs) at specific genomic loci. By contrast, CRISPR/Cas9 is adapted from a bacterial type II adaptive immune system, which uses a guide RNA and Cas9 nuclease to target specific genes for removal, replacement, or alteration [94]. A significant challenge in gene therapy is the limitation of off-target effects. For example, the zinc-finger protein ZNF274 has been implicated in silencing maternal SNORD116 and can activate its expression in PWS-induced pluripotent stem cells (iPSCs) when inhibited, although it has many additional targets in addition to SNORD116. iPSCs and their derived neurons from PWS patients with a variety of deletion sizes have been shown to retain the molecular signatures of PWS, as confirmed by MS-MLPA and CGH [95]. As a potential method of limiting off-target effects, Lagnouet et al. [92] demonstrate the feasibility of using CRISPR/Cas9 to delete specific ZNF274 binding sites at SNORD116 to limit its effects to that locus, which increased expression in PWS iPSCs that had been differentiated into forebrain cortical neurons. Such an approach is likely safer and more clinically feasible than the alteration of genome-wide epigenetic regulators. Additional targets for gene therapy under current investigation include the removal of SMCHD1, which encodes a protein implicated in the silencing of maternal PWS genes [96], as well as single-dose gene replacement of BDNF in the PWS mouse model, which has been previously shown to have a therapeutic effect in the treatment of obesity in non-PWS animal models [97].
Epigenetic therapies in contrast involve mechanisms of altering genetic expression that do not directly change a patient’s genetic sequence. Kim et al. [44] explored the use of two novel small molecules (▣UNC0642 and UNC0638) as epigenetic therapies in a PWS mouse model. The human 15q11-q13 imprinting region is highly conserved in mice, although without a deletion homologous to the humane type I and II deletions. The PWS mice in this study exhibited hyperphagia but not the obesity phenotype. The inhibitors of EMHT2/G9a (histone 3 lysine 9 methyltransferase) were capable of reactivating SNRPN and SNORD116, both in the mouse model and in human patient-derived fibroblasts in vitro. Earlier studies by Kim et al. [98] demonstrated a survival benefit in PWS model mouse pups using the same small molecule EMHT2/G9a inhibitors. Off-target effects are of significant concern, but the drug was well tolerated in the mouse model in both studies with no signs of toxicity and mirrors FD-approved epigenetic drugs used in cancer treatments, such as the DNA methyltransferase inhibitor decitabine, which is approved for the treatment of myelodysplastic syndromes and has been investigated as a potential therapy for PWS [45,98,99]. However, many questions regarding the cell types to be targeted, stages of development at which therapy should be initiated, precisely which genes need to be targeted, and an acceptable level for off-target effects must be answered before genetic and epigenetic therapies are investigated in PWS patients in vivo [100].
10. Conclusions
PWS is a complex, heterogeneous disease that has a significant impact on the lives of both the affected individuals and their families. There have been a significant number of advances in our understanding of the genetic mechanisms underlying the disease, and there are promising new discoveries on the horizon. However, the challenge of early diagnosis and difficulty in treating some of the disease’s most disabling features remain. As our understanding of PWS continues to grow, so does the availability of new therapies and management strategies available to clinicians and families.
Author Contributions
(I) Conception and design: all authors; (II) administrative support: none; (III) provision of study materials or patients: none; (IV) collection and assembly of data: all authors; (V) data analysis and interpretation: all authors; (VI) manuscript writing: all authors; (VII) final approval of manuscript: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
All authors declare no conflict of interest.
Abbreviations
| BDNF | Brain-derived neurotrophic factor |
| BMI | Body mass index |
| BP | Break points |
| CGH | Comparative genetic hybridization |
| CLOCK:BMAL | Circadian locomotor output cycles kaput brain and muscle ARNT-Like 1 |
| CMA | Chromosomal microarray analysis |
| SNURF | SNRPN upstream open reading frame |
| CPP | Central precocious puberty |
| CR1 | Complement C3b/C4b receptor 1 |
| CRY1/2 | Cryptochrome circadian regulator 1/2 |
| CVS | Chorionic villus sampling |
| CYFIP1 | CytoplasmicFMR1 interacting protein 1 |
| CYP | Cytochrome P450 |
| EMHT2/G9a | Euchromatic histone lysine methyltransferase 2 |
| FISH | Fluorescence in situ hybridization |
| GH | Growth hormone |
| GLP-1 | Glucagon-like peptide 1 |
| HPG | Hypothalamic-pituitary-gonadal |
| ICR | Imprinting control region |
| IGF-1 | Insulin-like growth factor 1 |
| KISS1 | Kisspeptin 1 |
| MAGEL2:MAGE | Family member L2 |
| MKRN3 | Makorin ring finger protein 3 |
| MS-MLPA | Methylation-specific multiplex ligation-dependent probe amplification |
| MS-PCR | Methylation-specific polymerase chain reaction |
| mUPD | Maternal uniparental disomy |
| NIPA1/2 | NIPA magnesium transporter 1/2 |
| NIPS | Non-invasive prenatal screening |
| OCD | Obsessive-compulsive disorder |
| OMIM | Online Mendelian Inheritance In Man |
| PCR | Polymerase chain reaction |
| PER | Period circadian protein homolog |
| PWS | Prader–Willi syndrome |
| rhGH | Recombinant human growth hormone |
| SCF | Skp, Cullin, F-box containing coplex |
| SMCHD1 | Structural Maintenance of Chromosomes flexible Hinge Domain Containing 1 |
| SNORD115/116 | Small nucleolar RNA, C/D box 115/116 cluster |
| SNP | single nucleotide polymorphisms |
| SNURPN | Small nucleolar ribonucleoprotein polypeptide N |
| SSRI | Selective serotonin reuptake inhibitor |
| STR | Short tandem repeat |
| TAC3 | Tachykinin precursor 3 |
| TUBGCP5 | Tubulin gamma complex associated protein 5 |
| UBE3a | Ubiquitin protein ligase E3A |
| USP7 | Ubiquitin-specific-processing protease 7 |
References
- Down, J.L. Lettsomian Lectures on some of the Mental Affections of Childhood and Youth. Br. Med. J. 1887, 1, 149–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prader, A.; Labhart, A.; Willi, H. Ein Syndrome von Adipositas, Kleinwuchs, Kryptochismus und Oligophrenie nach myatonieartigem Zustand in Neugeborenenalter. Schweiz Med. Wochenschr 1956, 86, 1260–1261. [Google Scholar]
- Butler, M.G.; Miller, J.L.; Forster, J.L. Prader-Willi Syndrome-Clinical Genetics, Diagnosis and Treatment Approaches: An Update. Curr. Pediatr. Rev. 2019, 15, 207–244. [Google Scholar] [CrossRef]
- Butler, M.G. Prader-Willi syndrome: Current understanding of cause and diagnosis. Am. J. Med. Genet. 1990, 35, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Gross, N.; Rabinowitz, R.; Gross-Tsur, V.; Hirsch, H.J.; Eldar-Geva, T. Prader-Willi syndrome can be diagnosed prenatally. Am. J. Med. Genet. Part A 2015, 167, 80–85. [Google Scholar] [CrossRef]
- Cassidy, S.B.; Schwartz, S.; Miller, J.L.; Driscoll, D.J. Prader-Willi syndrome. Genet. Med. 2012, 14, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G.; Manzardo, A.M.; Heinemann, J.; Loker, C.; Loker, J. Causes of death in Prader-Willi syndrome: Prader-Willi Syndrome Association (USA) 40-year mortality survey. Genet. Med. 2017, 19, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, S.B.; Driscoll, D.J. Prader-Willi syndrome. Eur. J. Hum. Genet. 2009, 17, 3–13. [Google Scholar] [CrossRef]
- Angulo, M.A.; Butler, M.G.; Cataletto, M.E. Prader-Willi syndrome: A review of clinical, genetic, and endocrine findings. J. Endocrinol. Investig. 2015, 38, 1249–1263. [Google Scholar] [CrossRef]
- Kalsner, L.; Chamberlain, S.J. Prader-Willi, Angelman, and 15q11-q13 Duplication Syndromes. Pediatr. Clin. N. Am. 2015, 62, 587–606. [Google Scholar] [CrossRef]
- Cattanach, B.M.; Kirk, M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 1985, 315, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G.; Hartin, S.N.; Hossain, W.A.; Manzardo, A.M.; Kimonis, V.; Dykens, E.; Gold, J.A.; Kim, S.-J.; Weisensel, N.; Tamura, R.; et al. Molecular genetic classification in Prader-Willi syndrome: A multisite cohort study. J. Med. Genet. 2018, 56, 149–153. [Google Scholar] [CrossRef]
- Lu, R.; Dong, Y.; Li, J.-D. Necdin regulates BMAL1 stability and circadian clock through SGT1-HSP90 chaperone machinery. Nucleic Acids Res. 2020, 48, 7944–7957. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.M.; Butler, M.G. The 15q11.2 BP1-BP2 microdeletion syndrome: A review. Int. J. Mol. Sci. 2015, 16, 4068–4082. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.-H.; Locke, D.; Greally, J.; Knoll, J.; Ohta, T.; Dunai, J.; Yavor, A.; Eichler, E.; Nicholls, R. Identification of Four Highly Conserved Genes between Breakpoint Hotspots BP1 and BP2 of the Prader-Willi/Angelman Syndromes Deletion Region That Have Undergone Evolutionary Transposition Mediated by Flanking Duplicons. Am. J. Hum. Genet. 2003, 73, 898–925. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G.; Fischer, W.; Kibiryeva, N.; Bittel, D.C. Array comparative genomic hybridization (aCGH) analysis in Prader-Willi syndrome. Am. J. Med Genet. Part A 2008, 146A, 854–860. [Google Scholar] [CrossRef]
- Butler, M.G.; Bittel, D.C.; Kibiryeva, N.; Talebizadeh, Z.; Thompson, T. Behavioral Differences Among Subjects with Prader-Willi Syndrome and Type I or Type II Deletion and Maternal Disomy. Pediatrics 2004, 113, 565–573. [Google Scholar] [CrossRef]
- Bittel, D.C.; Kibiryeva, N.; Butler, M.G. Expression of 4 Genes Between Chromosome 15 Breakpoints 1 and 2 and Behavioral Outcomes in Prader-Willi Syndrome. Pediatrics 2006, 118, e1276-83. [Google Scholar] [CrossRef]
- Murthy, S.; Nygren, A.; El Shakankiry, H.; Schouten, J.; Al Khayat, A.; Ridha, A.; Al Ali, M. Detection of a novel familial deletion of four genes between BP1 and BP2 of the Prader-Willi/Angelman syndrome critical region by oligo-array CGH in a child with neurological disorder and speech impairment. Cytogenet. Genome Res. 2007, 116, 135–140. [Google Scholar] [CrossRef]
- Doornbos, M.; Sikkema-Raddatz, B.; Ruijvenkamp, C.A.; Dijkhuizen, T.; Bijlsma, E.K.; Gijsbers, A.C.; Hilhorst-Hofstee, Y.; Hordijk, R.; Verbruggen, K.T.; Kerstjens-Frederikse, W.; et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader–Willi critical region, possibly associated with behavioural disturbances. Eur. J. Med Genet. 2009, 52, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.P.; Dauber, A.; Macedo, D.B.; Noel, S.D.; Brito, V.N.; Gill, J.C.; Cukier, P.; Thompson, I.R.; Navarro, V.M.; Gagliardi, P.C.; et al. Central Precocious Puberty Caused by Mutations in the Imprinted Gene MKRN3. N. Engl. J. Med. 2013, 368, 2467–2475. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.P.; Toro, C.A.; Song, Y.B.; Navarro, V.M.; Bosch, M.A.; Eren, A.; Liang, J.N.; Carroll, R.S.; Latronico, A.C.; Ronnekleiv, O.K.; et al. MKRN3 inhibits the reproductive axis through actions in kisspeptin-expressing neurons. J. Clin. Investig. 2020, 130, 4486–4500. [Google Scholar] [CrossRef]
- Carias, K.V.; Zoeteman, M.; Seewald, A.; Sanderson, M.R.; Bischof, J.M.; Wevrick, R. A MAGEL2-deubiquitinase complex modulates the ubiquitination of circadian rhythm protein CRY1. PLoS ONE 2020, 15, e0230874. [Google Scholar] [CrossRef] [PubMed]
- Tacer, K.F.; Potts, P.R. Cellular and disease functions of the Prader–Willi Syndrome gene MAGEL2. Biochem. J. 2017, 474, 2177–2190. [Google Scholar] [CrossRef]
- Schaaf, C.P.; Gonzalez-Garay, M.L.; Xia, F.; Potocki, L.; Gripp, K.W.; Zhang, B.; Peters, B.A.; A McElwain, M.; Drmanac, R.; Beaudet, A.L.; et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat. Genet. 2013, 45, 1405–1408. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, T.; Del Gaudio, D.; German, J.R.; Shinawi, M.; Peters, S.U.; Person, R.E.; Garnica, A.; Cheung, S.W.; Beaudet, A.L. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008, 40, 719–721. [Google Scholar] [CrossRef]
- De Smith, A.J.; Purmann, C.; Walters, R.; Ellis, R.J.; Holder, S.E.; Van Haelst, M.M.; Brady, A.F.; Fairbrother, U.L.; Dattani, M.; Keogh, J.M.; et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 2009, 18, 3257–3265. [Google Scholar] [CrossRef]
- Yin, Q.-F.; Yang, L.; Zhang, Y.; Xiang, J.-F.; Wu, Y.-W.; Carmichael, G.G.; Chen, L.-L. Long Noncoding RNAs with snoRNA Ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef]
- Coulson, R.L.; Yasui, D.H.; Dunaway, K.W.; Laufer, B.I.; Ciernia, A.V.; Zhu, Y.; Mordaunt, C.E.; Totah, T.S.; LaSalle, J.M. Snord116-dependent diurnal rhythm of DNA methylation in mouse cortex. Nat. Commun. 2018, 9, 1616. [Google Scholar] [CrossRef]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-H.; Mohawk, J.A.; Siepka, S.M.; Shan, Y.; Huh, S.K.; Hong, H.-K.; Kornblum, I.; Kumar, V.; Koike, N.; Xu, M.; et al. Competing E3 Ubiquitin Ligases Govern Circadian Periodicity by Degradation of CRY in Nucleus and Cytoplasm. Cell 2013, 152, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Runte, M.; Varon, R.; Horn, D.; Horsthemke, B.; Buiting, K. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Qual. Life Res. 2004, 116, 228–230. [Google Scholar] [CrossRef]
- Ledbetter, D.H.; Riccardi, V.M.; Airhart, S.D.; Strobel, R.J.; Keenan, B.S.; Crawford, J.D. Deletions of Chromosome 15 as a Cause of the Prader–Willi Syndrome. N. Engl. J. Med. 1981, 304, 325–329. [Google Scholar] [CrossRef]
- Butler, M.G. Benefits and limitations of prenatal screening for Prader-Willi syndrome. Prenat Diagn. 2017, 37, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Kimonis, V.E.; Tamura, R.; Gold, J.-A.; Patel, N.; Surampalli, A.; Manazir, J.; Miller, J.L.; Roof, E.; Dykens, E.; Butler, M.G.; et al. Early Diagnosis in Prader–Willi Syndrome Reduces Obesity and Associated Co-Morbidities. Genes 2019, 10, 898. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, X.; Wang, J.; Zhang, Y.; Wang, A.; Lu, J.; Huang, Y.; Liu, S.; Wu, J.; Du, L.; et al. Genetic testing for Prader-Willi syndrome and Angelman syndrome in the clinical practice of Guangdong Province, China. Mol. Cytogenet. 2019, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Srebnik, N.; Even-Zohar, N.G.; Salama, A.; Sela, H.Y.; Hirsch, H.J.; Gross-Tsur, V.; Eldar-Geva, T. Recognizing the unique prenatal phenotype of Prader-Willi Syndrome (PWS) indicates the need for a diagnostic methylation test. Prenat. Diagn. 2020, 40, 878–884. [Google Scholar] [CrossRef]
- Yang, L.; Ma, B.; Mao, S.; Zhou, Q.; Zou, C. Establishing perinatal and neonatal features of Prader-Willi syndrome for efficient diagnosis and outcomes. Expert Opin. Orphan Drugs 2020, 8, 265–271. [Google Scholar] [CrossRef]
- Chen, C.; Visootsak, J.; Dills, S.; Graham, J.M. Prader-Willi Syndrome: An Update and Review for the Primary Pediatrician. Clin. Pediatr. 2007, 46, 580–591. [Google Scholar] [CrossRef]
- Strom, S.P.; Hossain, W.A.; Grigorian, M.; Li, M.; Fierro, J.; Scaringe, W.; Yen, H.-Y.; Teguh, M.; Liu, J.; Gao, H.; et al. A Streamlined Approach to Prader-Willi and Angelman Syndrome Molecular Diagnostics. Front. Genet. 2021, 12, 608889. [Google Scholar] [CrossRef]
- Rosenberg, A.G.W.; Pater, M.R.A.; Pellikaan, K.; Davidse, K.; Kattentidt-Mouravieva, A.A.; Kersseboom, R.; Bos-Roubos, A.G.; van Eeghen, A.; Veen, J.M.C.; van der Meulen, J.J.; et al. What Every Internist-Endocrinologist Should Know about Rare Genetic Syndromes in Order to Prevent Needless Diagnostics, Missed Diagnoses and Medical Complications: Five Years of ‘Internal Medicine for Rare Genetic Syndromes’. J. Clin. Med. 2021, 10, 5457. [Google Scholar] [CrossRef] [PubMed]
- Holm, V.A.; Cassidy, S.B.; Butler, M.G.; Hanchett, J.M.; Greenswag, L.R.; Whitman, B.Y.; Greenberg, F. Prader-Willi Syndrome: Consensus Diagnostic Criteria. Pediatrics 1993, 91, 398–402. [Google Scholar] [CrossRef]
- Kim, Y.; Wang, S.E.; Jiang, Y.-H. Epigenetic therapy of Prader–Willi syndrome. Transl. Res. 2019, 208, 105–118. [Google Scholar] [CrossRef] [PubMed]
- A Méndez-Rosado, L.; García, D.; Molina-Gamboa, O.; García, A.; De León, N.; Lantigua-Cruz, A.; Liehr, T. Algorithm for the diagnosis of patients with neurodevelopmental disorders and suspicion of a genetic syndrome. Arch. Argent. Pediatr. 2020, 118, 52–55. [Google Scholar] [CrossRef]
- Gunay-Aygun, M.; Schwartz, S.; Heeger, S.; O’Riordan, M.A.; Cassidy, S.B. The Changing Purpose of Prader-Willi Syndrome Clinical Diagnostic Criteria and Proposed Revised Criteria. Pediatrics 2001, 108, e92. [Google Scholar] [CrossRef]
- El-Bassyouni, H.T.; Hassan, N.; Mahfouz, I.; Abd-Elnaby, A.E.; Mostafa, M.I.; Tosson, A.M. Early Detection and Management of Prader-Willi Syndrome in Egyptian Patients. J. Pediatr. Genet. 2019, 08, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Traisrisilp, K.; Sirikunalai, P.; Sirilert, S.; Chareonsirisuthigul, T.; Tongsong, T. Cardiac rhabdomyoma as a possible new prenatal sonographic feature of Prader–Willi syndrome. J. Obstet. Gynaecol. Res. 2021, 48, 239–243. [Google Scholar] [CrossRef]
- Monaghan, K.G.; Wiktor, A.; Van Dyke, D.L. Diagnostic testing for Prader-Willi syndrome and Angelman syndrome: A cost comparison. Genet. Med. 2002, 4, 448–450. [Google Scholar] [CrossRef] [PubMed]
- Tauber, M.; Hoybye, C. Endocrine disorders in Prader-Willi syndrome: A model to understand and treat hypothalamic dysfunction. Lancet Diabetes Endocrinol. 2021, 9, 235–246. [Google Scholar] [CrossRef]
- Beygo, J.; Buiting, K.; Ramsden, S.C.; Ellis, R.; Clayton-Smith, J.; Kanber, D. Update of the EMQN/ACGS best practice guidelines for molecular analysis of Prader-Willi and Angelman syndromes. Eur. J. Hum. Genet. 2019, 27, 1326–1340. [Google Scholar] [CrossRef]
- Dong, Y.; Liu, S.; Li, J.; Li, J.; Chen, Q.; Luo, J.; Li, C.; Li, H.; Qi, H.; Li, R. Possibility of early diagnosis in a fetus affected by Prader-Willi syndrome with maternal hetero-UPD15: A lesson to be learned. Mol. Med. Rep. 2019, 20, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Pellikaan, K.; Rosenberg, A.G.W.; Kattentidt-Mouravieva, A.A.; Kersseboom, R.; Bos-Roubos, A.G.; Veen-Roelofs, J.M.C.; Wieringen, N.V.A.-V.; Hoekstra, F.M.E.; Berg, S.A.A.V.D.; Van Der Lely, A.J.; et al. Missed Diagnoses and Health Problems in Adults with Prader-Willi Syndrome: Recommendations for Screening and Treatment. J. Clin. Endocrinol. Metab. 2020, 105, e4671–e4687. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, S.; Feng, B.; Yang, Y.; Zhang, H.; Dong, R.; Liu, Y.; Gai, Z. Clinical Application of an Innovative Multiplex-Fluorescent-Labeled STRs Assay for Prader-Willi Syndrome and Angelman Syndrome. PLoS ONE 2016, 11, e0147824. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Park, Y.; Cho, S.I.; Kim, M.J.; Chae, J.-H.; Kim, J.Y.; Seong, M.-W.; Park, A.S.S. Clinical Utility of Methylation-Specific Multiplex Ligation-Dependent Probe Amplification for the Diagnosis of Prader–Willi Syndrome and Angelman Syndrome. Ann. Lab. Med. 2022, 42, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Law, H.-Y.; Chong, S.S. Detection and Discrimination between Deletional and Non-Deletional Prader-Willi and Angelman Syndromes by Methylation-Specific PCR and Quantitative Melting Curve Analysis. J. Mol. Diagn. 2009, 11, 446–449. [Google Scholar] [CrossRef][Green Version]
- AlBanji, M.H.; AlSaad, A.N.; AlAnazi, R.F.; Aleisa, Z.A.; Alam, D.S.; Alhashim, A.H. Utility of Hypotonia Diagnostic Investigations: A 12-year Single Center Study. Mol. Genet. Metab. Rep. 2020, 25, 100665. [Google Scholar] [CrossRef]
- Li, H.; Meng, S.; Chen, Z.; Li, H.; Du, M.; Ma, H.; Wei, H.; Duan, H.; Zheng, H.; Wenren, Q.; et al. Molecular Genetic Diagnostics of Prader-Willi Syndrome: A Validation of Linkage Analysis for the Chinese Population. J. Genet. Genom. 2007, 34, 885–891. [Google Scholar] [CrossRef]
- Driscoll, D.; Miller, J.; Schwartz, S.; Cassidy, S. Prader-Willi Syndrome. Neuropediatrics 2017, 29, 124–126. [Google Scholar]
- Yang, L.; Zhou, Q.; Ma, B.; Mao, S.; Dai, Y.; Zhu, M.; Zou, C. Perinatal features of Prader-Willi syndrome: A Chinese cohort of 134 patients. Orphanet J. Rare Dis. 2020, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Manning, F.A. FETAL BIOPHYSICAL PROFILE. Obstet. Gynecol. Clin. N. Am. 1999, 26, 557. [Google Scholar] [CrossRef]
- Cassidy, S.B.; Forsythe, M.; Heeger, S.; Nicholls, R.D.; Schork, N.; Benn, P.; Schwartz, S. Comparison of phenotype between patients with Prader-Willi syndrome due to deletion 15q and uniparental disomy 15. Am. J. Med. Genet. 1997, 68, 433–440. [Google Scholar] [CrossRef]
- Bamfo, J.E.A.K.; Odibo, A.O. Diagnosis and Management of Fetal Growth Restriction. J. Pregnancy 2011, 2011, 640715. [Google Scholar] [CrossRef]
- Shubina, J.; Barkov, I.Y.; Stupko, O.K.; Kuznetsova, M.V.; Goltsov, A.Y.; Kochetkova, T.O.; Trofimov, D.Y.; Sukhikh, G.T. Prenatal diagnosis of Prader-Willi syndrome due to uniparental disomy with NIPS: Case report and literature review. Mol. Genet. Genom. Med. 2020, 8, e1448. [Google Scholar] [CrossRef]
- Gross, N.E.-Z.; Geva-Eldar, T.; Pollak, Y.; Hirsch, H.J.; Gross, I.; Gross-Tsur, V. Attitudes toward prenatal genetic testing and therapeutic termination of pregnancy among parents of offspring with Prader-Willi syndrome. Eur. J. Med Genet. 2017, 60, 205–211. [Google Scholar] [CrossRef]
- Cobo, J.; Coronas, R.; Pousa, E.; Oliva, J.-C.; Giménez-Palop, O.; Esteba-Castillo, S.; Novell, R.; Palao, D.; Caixàs, A. Multidimensional Evaluation of Awareness in Prader-Willi Syndrome. J. Clin. Med. 2021, 10, 2007. [Google Scholar] [CrossRef] [PubMed]
- Heksch, R.; Kamboj, M.; Anglin, K.; Obrynba, K. Review of Prader-Willi syndrome: The endocrine approach. Transl. Pediatr. 2017, 6, 274–285. [Google Scholar] [CrossRef]
- Whittington, J.E.; Holland, A.J.; Webb, T.; Butler, J.; Clarke, D.; Boer, H. Population prevalence and estimated birth incidence and mortality rate for people with Prader-Willi syndrome in one UK Health Region. J. Med. Genet. 2001, 38, 792–798. [Google Scholar] [CrossRef]
- Alfaro, D.L.P.; Lemoine, P.; Ehlinger, V.; Molinas, C.; Diene, G.; Valette, M.; Pinto, G.; Coupaye, M.; Poitou-Bernert, C.; Thuilleaux, D.; et al. Causes of death in Prader-Willi syndrome: Lessons from 11 years’ experience of a national reference center. Orphanet J. Rare Dis. 2019, 14, 238. [Google Scholar] [CrossRef] [PubMed]
- Höybye, C.; Holland, A.J.; Driscoll, D.J. Time for a general approval of growth hormone treatment in adults with Prader-Willi syndrome. Orphanet J. Rare Dis. 2021, 16, 69. [Google Scholar] [CrossRef]
- Deal, C.L.; Tony, M.; Höybye, C.; Allen, D.B.; Tauber, M.; Christiansen, J.S. GrowthHormone Research Society workshop summary: Consensus guidelines for recombinant human growth hormone therapy in Prader-Willi syndrome. J. Clin. Endocrinol. Metab. 2013, 98, E1072–E1087. [Google Scholar] [CrossRef] [PubMed]
- Lecka-Ambroziak, A.; Wysocka-Mincewicz, M.; Doleżal-Ołtarzewska, K.; Zygmunt-Górska, A.; Wędrychowicz, A.; Żak, T.; Noczyńska, A.; Birkholz-Walerzak, D.; Stawerska, R.; Hilczer, M.; et al. Effects of Recombinant Human Growth Hormone Treatment, Depending on the Therapy Start in Different Nutritional Phases in Paediatric Patients with Prader–Willi Syndrome: A Polish Multicentre Study. J. Clin. Med. 2021, 10, 3176. [Google Scholar] [CrossRef]
- Donze, S.H.; Damen, L.; Mahabier, E.F.; Hokken-Koelega, A.C. Cognitive functioning in children with Prader–Willi syndrome during 8 years of growth hormone treatment. Eur. J. Endocrinol. 2020, 182, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Angulo, M.; Abuzzahab, M.J.; Pietropoli, A.; Ostrow, V.; Kelepouris, N.; Tauber, M. Outcomes in children treated with growth hormone for Prader-Willi syndrome: Data from the ANSWER Program® and NordiNet® International Outcome Study. Int. J. Pediatr. Endocrinol. 2020, 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Höybye, C.; Thorén, M.; Böhm, B. Cognitive, emotional, physical and social effects of growth hormone treatment in adults with Prader-Willi syndrome. J. Intellect. Disabil. Res. 2005, 49, 245–252. [Google Scholar] [CrossRef]
- Donze, S.H.; Kuppens, R.J.; Bakker, N.E.; Velden, J.A.V.A.-V.D.; Hokken-Koelega, A.C. Bone mineral density in young adults with Prader-Willi syndrome: A randomized, placebo-controlled, crossover GH trial. Clin. Endocrinol. 2018, 88, 806–812. [Google Scholar] [CrossRef]
- Oto, Y.; Murakami, N.; Matsubara, K.; Saima, S.; Ogata, H.; Ihara, H.; Nagai, T.; Matsubara, T. Effects of growth hormone treatment on thyroid function in pediatric patients with Prader–Willi syndrome. Am. J. Med Genet. Part A 2020, 182, 659–663. [Google Scholar] [CrossRef]
- Tan, Q.; Orsso, C.E.; Deehan, E.C.; Triador, L.; Field, C.; Tun, H.M.; Han, J.C.; Müller, T.D.; Haqq, A.M. Current and emerging therapies for managing hyperphagia and obesity in Prader-Willi syndrome: A narrative review. Obes. Rev. 2019, 21, e12992. [Google Scholar] [CrossRef]
- Michel, L.M.; Haqq, A.M.; Wismer, W.V. A review of chemosensory perceptions, food preferences and food-related behaviours in subjects with Prader–Willi Syndrome. Appetite 2016, 99, 17–24. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Lee, Y.J.; Kim, S.Y.; Cheon, C.K.; Lim, H.H. Successful rapid weight reduction and the use of liraglutide for morbid obesity in adolescent Prader-Willi syndrome. Ann. Pediatr. Endocrinol. Metab. 2020, 25, 52–56. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Barrea, L.; Faggiano, F.; Maiorino, M.I.; Parrillo, M.; Pugliese, G.; Ruggeri, R.M.; Scarano, E.; Savastano, S.; Colao, A.; et al. Obesity in Prader–Willi syndrome: Physiopathological mechanisms, nutritional and pharmacological approaches. J. Endocrinol. Investig. 2021, 44, 2057–2070. [Google Scholar] [CrossRef] [PubMed]
- Consoli, A.; Berthoumieu, S.; Raffin, M.; Thuilleaux, D.; Poitou, C.; Coupaye, M.; Pinto, G.; Lebbah, S.; Zahr, N.; Tauber, M.; et al. Effect of topiramate on eating behaviours in Prader-Willi syndrome: TOPRADER double-blind randomised placebo-controlled study. Transl. Psychiatry 2019, 9, 274. [Google Scholar] [CrossRef]
- Griggs, J.L.; Mathai, M.L.; Sinnayah, P. Caralluma fimbriata extract activity involves the 5-HT2c receptor in PWS Snord116 deletion mouse model. Brain Behav. 2018, 8, e01102. [Google Scholar] [CrossRef] [PubMed]
- Griggs, J. Single-Case Study of Appetite Control in Prader-Willi Syndrome, Over 12-Years by the Indian Extract Caralluma fimbriata. Genes 2019, 10, 447. [Google Scholar] [CrossRef] [PubMed]
- Duje, B.; Nevena, K.; Katja, D.K.; Anita, U.; Igor, M.; Jurica, V. 208 Use of GLP-1 analog in a patient with prader-willi syndrome. Abstracts 2021, 106, A88. [Google Scholar] [CrossRef]
- Fintini, D.; Grugni, G.; Brufani, C.; Bocchini, S.; Cappa, M.; Crinò, A. Use of GLP-1 Receptor Agonists in Prader-Willi Syndrome: Report of Six Cases. Diabetes Care 2014, 37, e76–e77. [Google Scholar] [CrossRef]
- Forster, J.; Duis, J.; Butler, M. Pharmacogenetic Testing of Cytochrome P450 Drug Metabolizing Enzymes in a Case Series of Patients with Prader-Willi Syndrome. Genes 2021, 12, 152. [Google Scholar] [CrossRef]
- Takahashi, P.Y.; Ryu, E.; Pathak, J.; Jenkins, G.D.; Batzler, A.; Hathcock, M.A.; Black, J.L.; Olson, J.E.; Cerhan, J.R.; Bielinski, S.J. Increased risk of hospitalization for ultrarapid metabolizers of cytochrome P450 2D6. Pharm. Pers. Med. 2017, 10, 39–47. [Google Scholar] [CrossRef]
- Pennington, S.; Stutzman, D.; Sannar, E. Pitolisant in an Adolescent with Prader-Willi Syndrome. J. Pediatr. Pharmacol. Ther. 2021, 26, 405–4100. [Google Scholar] [CrossRef]
- Schaller, F.; Watrin, F.; Sturny, R.; Massacrier, A.; Szepetowski, P.; Muscatelli, F. A single postnatal injection of oxytocin rescues the lethal feeding behaviour in mouse newborns deficient for the imprinted Magel2 gene. Hum. Mol. Genet. 2010, 19, 4895–4905. [Google Scholar] [CrossRef] [PubMed]
- Damen, L.; Grootjen, L.N.; Juriaans, A.F.; Donze, S.H.; Huisman, T.M.; Visser, J.A.; Delhanty, P.J.; Hokken-Koelega, A.C. Oxytocin in young children with Prader-Willi syndrome: Results of a randomized, double-blind, placebo-controlled, crossover trial investigating 3 months of oxytocin. Clin. Endocrinol. 2020, 94, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Langouët, M.; Gorka, D.; Orniacki, C.; Dupont-Thibert, C.M.; Chung, M.S.; Glatt-Deeley, H.R.; Germain, N.; Crandall, L.J.; Cotney, J.L.; E Stoddard, C.; et al. Specific ZNF274 binding interference at SNORD116 activates the maternal transcripts in Prader-Willi syndrome neurons. Hum. Mol. Genet. 2020, 29, 3285–3295. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Syding, L.A.; Nickl, P.; Kasparek, P.; Sedlacek, R. CRISPR/Cas9 Epigenome Editing Potential for Rare Imprinting Diseases: A Review. Cells 2020, 9, 993. [Google Scholar] [CrossRef]
- Burnett, L.C.; LeDuc, C.A.; Sulsona, C.R.; Paull, D.; Eddiry, S.; Levy, B.; Salles, J.P.; Tauber, M.; Driscoll, D.J.; Egli, D.; et al. Induced pluripotent stem cells (iPSC) created from skin fibroblasts of patients with Prader-Willi syndrome (PWS) retain the molecular signature of PWS. Stem Cell Res. 2016, 17, 526–530. [Google Scholar] [CrossRef]
- Foundation for Prader-Willi Research. Targeting SMCHD1 to Address the Underlying Cause of PWS and SYS. Available online: https://www.fpwr.org/fpwr-funded-projects/targeting-smchd1-to-address-the-underlying-cause-of-pws-and-sys (accessed on 18 April 2022).
- Foundation for Prader-Willi Research. Gene Therapy of Obesity in Prader-Willi Syndrome by an Autoregulatory BDNF Vector. Available online: https://www.fpwr.org/fpwr-funded-projects/gene-therapy-of-obesity-in-prader-willi-syndrome-by-an-autoregulatory-bdnf-vector (accessed on 18 April 2022).
- Kim, Y.; Lee, H.-M.; Xiong, Y.; Sciaky, N.; Hulbert, S.W.; Cao, X.; Everitt, J.I.; Jin, J.; Roth, B.L.; Jiang, Y.-H. Targeting the histone methyltransferase G9a activates imprinted genes and improves survival of a mouse model of Prader–Willi syndrome. Nat. Med. 2016, 23, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Crunkhorn, S. Steps towards epigenetic therapy for PWS. Nat. Rev. Drug Discov. 2017, 16, 85. [Google Scholar] [CrossRef]
- Foundation for Prader-Willi Research. Genetic Therapy for Prader-Willi Syndrome. Available online: https://www.fpwr.org/genetic-therapy-for-prader-willi-syndrome (accessed on 18 April 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).