Pathway Analysis of Genome Wide Association Studies (GWAS) Data Associated with Male Infertility
Abstract
:1. Introduction
2. Methods
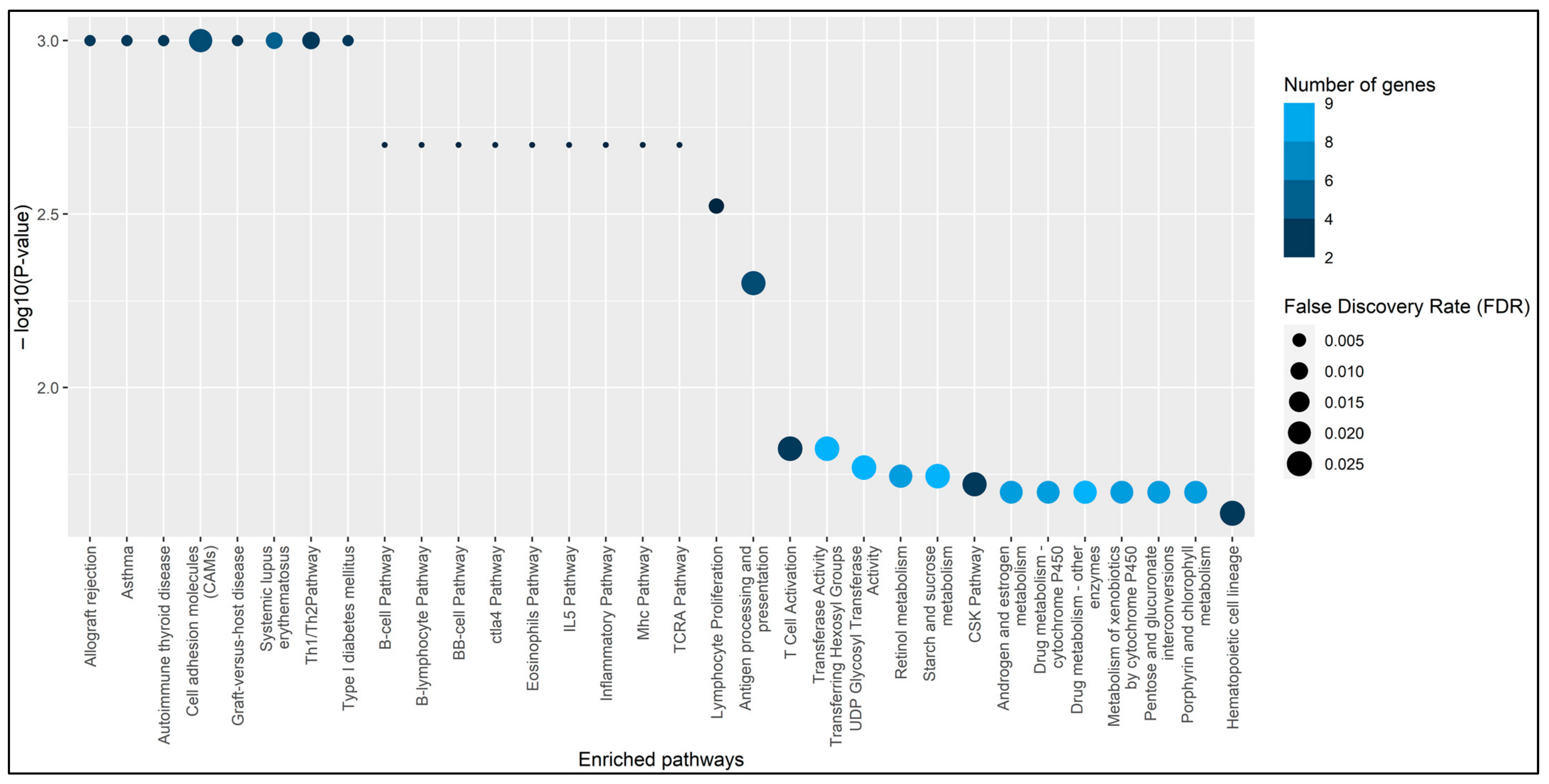
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Callahan, T.; Caughey, A.B. Infertility and Assisted Reproductive Technologies. In Blueprints Obstetrics and Gynecology; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008; Volume 5. [Google Scholar]
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A Unique View on Male Infertility around the Globe. Reprod. Biol. Endocrinol. 2015, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Poongothai, J.; Gopenath, T.S.; Manonayaki, S. Genetics of Human Male Infertility. Singap. Med. J. 2009, 50, 336–347. [Google Scholar]
- Singh, K.; Jaiswal, D. Human Male Infertility: A Complex Multifactorial Phenotype. Reprod. Sci. 2011, 18, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Karamzade, A.; Mirzapour, H.; Kheirollahi, M. Genetics Aspects of Male Infertility. J. Isfahan Med. Sch. 2013, 31, 1149–1162. [Google Scholar]
- Choy, J.T.; Eisenberg, M.L. Male Infertility as a Window to Health. Fertil. Steril. 2018, 110, 810–814. [Google Scholar] [CrossRef]
- Lettieri, G.; D’Agostino, G.; Mele, E.; Cardito, C.; Esposito, R.; Cimmino, A.; Giarra, A.; Trifuoggi, M.; Raimondo, S.; Notari, T.; et al. Discovery of the Involvement in DNA Oxidative Damage of Human Sperm Nuclear Basic Proteins of Healthy Young Men Living in Polluted Areas. Int. J. Mol. Sci. 2020, 21, 4198. [Google Scholar] [CrossRef]
- Panner Selvam, M.K.; Sengupta, P.; Agarwal, A. Sperm DNA Fragmentation and Male Infertility. In Genetics of Male Infertility; Springer International Publishing: Cham, The Switzerland, 2020; pp. 155–172. [Google Scholar] [CrossRef]
- Lettieri, G.; Marra, F.; Moriello, C.; Prisco, M.; Notari, T.; Trifuoggi, M.; Giarra, A.; Bosco, L.; Montano, L.; Piscopo, M. Molecular Alterations in Spermatozoa of a Family Case Living in the Land of Fires—A First Look at Possible Transgenerational Effects of Pollutants. Int. J. Mol. Sci. 2020, 21, 6710. [Google Scholar] [CrossRef]
- Sharpe, R.M. Environmental/Lifestyle Effects on Spermatogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1697–1712. [Google Scholar] [CrossRef]
- Kumar, N.; Singh, A. Trends of Male Factor Infertility, an Important Cause of Infertility: A Review of Literature. J. Hum. Reprod. Sci. 2015, 8, 191–196. [Google Scholar] [CrossRef]
- Sadeghi-Bazargani, H.; Hajshafiha; Ghareaghaji; Salemi; Sadeghi-asadi. Association of Body Mass Index with Some Fertility Markers among Male Partners of Infertile Couples. Int. J. Gen. Med. 2013, 6, 447–451. [Google Scholar] [CrossRef]
- Krausz, C.; Riera-Escamilla, A. Genetics of Male Infertility. Nat. Rev. Urol. 2018, 15, 369–384. [Google Scholar] [CrossRef] [PubMed]
- Dada, R.; Shamsi, M.; Kumar, K. Genetic and Epigenetic Factors: Role in Male Infertility. Indian J. Urol. 2011, 27, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.; Milne, S.; Leeson, H. Sperm DNA Damage Caused by Oxidative Stress: Modifiable Clinical, Lifestyle and Nutritional Factors in Male Infertility. Reprod. BioMedicine Online 2014, 28, 684–703. [Google Scholar] [CrossRef] [PubMed]
- Karam, Z.M.; Baba Salari, M.; Anjom Shoaa, A.; Dehghan Kouhestani, S.; Bahram Nejad, A.; Ashourzadeh, S.; Zangouyee, M.R.; Bazrafshani, M.R. Impact of Oxidative Stress SNPs on Sperm DNA Damage and Male Infertility in a South-East Iranian Population. Reprod. Fertil. Dev. 2022, 34, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Wouters-Tyrou, D.; Martinage, A.; Chevaillier, P.; Sautière, P. Nuclear Basic Proteins in Spermiogenesis. Biochimie 1998, 80, 117–128. [Google Scholar] [CrossRef]
- Sassone-Corsi, P. Unique Chromatin Remodeling and Transcriptional Regulation in Spermatogenesis. Science 2002, 296, 2176–2178. [Google Scholar] [CrossRef]
- Kimmins, S.; Sassone-Corsi, P. Chromatin Remodelling and Epigenetic Features of Germ Cells. Nature 2005, 434, 583–589. [Google Scholar] [CrossRef]
- Iguchi, N.; Yang, S.; Lamb, D.J.; Hecht, N.B. An SNP in Protamine 1: A Possible Genetic Cause of Male Infertility? J. Med. Genet. 2005, 43, 382–384. [Google Scholar] [CrossRef]
- Venkatesh, S.; Kumar, R.; Deka, D.; Deecaraman, M.; Dada, R. Analysis of Sperm Nuclear Protein Gene Polymorphisms and DNA Integrity in Infertile Men. Syst. Biol. Reprod. Med. 2011, 57, 124–132. [Google Scholar] [CrossRef]
- Guerrero-Bosagna, C.; Skinner, M.K. Environmentally Induced Epigenetic Transgenerational Inheritance of Male Infertility. Curr. Opin. Genet. Dev. 2014, 26, 79–88. [Google Scholar] [CrossRef]
- Raimondo, S.; Gentile, M.; Esposito, G.; Gentile, T.; Ferrara, I.; Crescenzo, C.; Palmieri, M.; Cuomo, F.; de Filippo, S.; Lettieri, G.; et al. Could Kallikrein-Related Serine Peptidase 3 Be an Early Biomarker of Environmental Exposure in Young Women? Int. J. Environ. Res. Public Health 2021, 18, 8833. [Google Scholar] [CrossRef] [PubMed]
- Ferlin, A.; Arredi, B.; Foresta, C. Genetic Causes of Male Infertility. Reprod. Toxicol. 2006, 22, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Erenpreiss, J.; Elzanaty, S.; Giwercman, A. Sperm DNA Damage in Men from Infertile Couples. Asian J. Androl. 2008, 10, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Oleszczuk, K.; Giwercman, A.; Bungum, M. Intra-Individual Variation of the Sperm Chromatin Structure Assay DNA Fragmentation Index in Men from Infertile Couples. Hum. Reprod. 2011, 26, 3244–3248. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Long, Y.; Zhou, Y.; Huang, C.; Gu, A.; Wang, X. Common Variants in Mismatch Repair Genes Associated with Increased Risk of Sperm DNA Damage and Male Infertility. BMC Med. 2012, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Tsujimura, A.; Miyagawa, Y.; Koh, E.; Namiki, M.; Sengoku, K. Male Infertility and Its Causes in Human. Adv. Urol. 2012, 2012, 384520. [Google Scholar] [CrossRef]
- Krausz, C.; Giachini, C. Genetic Risk Factors in Male Infertility. Arch. Androl. 2007, 53, 125–133. [Google Scholar] [CrossRef]
- Lindgren, K.E.; Nordqvist, S.; Kårehed, K.; Sundström-Poromaa, I.; Åkerud, H. The Effect of a Specific Histidine-Rich Glycoprotein Polymorphism on Male Infertility and Semen Parameters. Reprod. BioMedicine Online 2016, 33, 180–188. [Google Scholar] [CrossRef]
- Schulte, R.T.; Ohl, D.A.; Sigman, M.; Smith, G.D. Sperm DNA Damage in Male Infertility: Etiologies, Assays, and Outcomes. J. Assist. Reprod. Genet. 2010, 27, 3–12. [Google Scholar] [CrossRef]
- Morris, I.D. The Spectrum of DNA Damage in Human Sperm Assessed by Single Cell Gel Electrophoresis (Comet Assay) and Its Relationship to Fertilization and Embryo Development. Hum. Reprod. 2002, 17, 990–998. [Google Scholar] [CrossRef]
- Gandini, L.; Lombardo, F.; Paoli, D.; Caruso, F.; Eleuteri, P.; Leter, G.; Ciriminna, R.; Culasso, F.; Dondero, F.; Lenzi, A.; et al. Full-Term Pregnancies Achieved with ICSI despite High Levels of Sperm Chromatin Damage. Hum. Reprod. 2004, 19, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, L.; Cai, J.; Huang, H. Correlation of Sperm DNA Damage with IVF and ICSI Outcomes: A Systematic Review and Meta-Analysis. J. Assist. Reprod. Genet. 2006, 23, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Benchaib, M. Sperm DNA Fragmentation Decreases the Pregnancy Rate in an Assisted Reproductive Technique. Hum. Reprod. 2003, 18, 1023–1028. [Google Scholar] [CrossRef]
- Larson, K.L. Sperm Chromatin Structure Assay Parameters as Predictors of Failed Pregnancy Following Assisted Reproductive Techniques. Hum. Reprod. 2000, 15, 1717–1722. [Google Scholar] [CrossRef]
- Virro, M.R.; Larson-Cook, K.L.; Evenson, D.P. Sperm Chromatin Structure Assay (Scsa®) Parameters Are Related to Fertilization, Blastocyst Development, and Ongoing Pregnancy in in Vitro Fertilization and Intracytoplasmic Sperm Injection Cycles. Fertil. Steril. 2004, 81, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Brunborg, G.; Stevenson, M.; Lutton, D.; McManus, J.; Lewis, S.E.M. Clinical Significance of Sperm DNA Damage in Assisted Reproduction Outcome. Hum. Reprod. 2010, 25, 1594–1608. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Li, Z.; Yu, J.; Tong, C.; Lin, Y.; Guo, X.; Lu, F.; Dong, J.; Xia, Y.; Wen, Y.; et al. Association Analysis Identifies New Risk Loci for Non-Obstructive Azoospermia in Chinese Men. Nat. Commun. 2014, 5, 3857. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, J.-H.; Song, G.G. Pathway Analysis of a Genome-Wide Association Study in Schizophrenia. Gene 2013, 525, 107–115. [Google Scholar] [CrossRef]
- Mortezaei, Z.; Tavallaei, M. Recent Innovations and In-Depth Aspects of Post-Genome Wide Association Study (Post-GWAS) to Understand the Genetic Basis of Complex Phenotypes. Heredity 2021, 127, 485–497. [Google Scholar] [CrossRef]
- Jia, P.; Wang, L.; Meltzer, H.Y.; Zhao, Z. Pathway-Based Analysis of GWAS Datasets: Effective but Caution Required. Int. J. Neuropsychopharmacol. 2011, 14, 567–572. [Google Scholar] [CrossRef]
- Yoon, S.; Nguyen, H.C.T.; Yoo, Y.J.; Kim, J.; Baik, B.; Kim, S.; Kim, J.; Kim, S.; Nam, D. Efficient Pathway Enrichment and Network Analysis of GWAS Summary Data Using GSA-SNP2. Nucleic Acids Res. 2018, 46, e60. [Google Scholar] [CrossRef] [PubMed]
- Matern, B.M.; Olieslagers, T.I.; Voorter, C.E.M.; Groeneweg, M.; Tilanus, M.G.J. Insights into the Polymorphism in HLA—DRA and Its Evolutionary Relationship with HLA Haplotypes. HLA 2020, 95, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Mosaad, Y.M. Clinical Role of Human Leukocyte Antigen in Health and Disease. Scand. J. Immunol. 2015, 82, 283–306. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.R.; Sun, D.; Freeman, W.M.; Lazarus, P. Quantification of Hepatic UDP Glucuronosyltransferase 1A Splice Variant Expression and Correlation of UDP Glucuronosyltransferase 1A1 Variant Expression with Glucuronidation Activity. J. Pharmacol. Exp. Ther. 2012, 342, 720–729. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Z.; Chang, W.; Liu, F.; Xie, J.; Yang, Y.; Qiu, H. Classification of Patients With Sepsis According to Immune Cell Characteristics: A Bioinformatic Analysis of Two Cohort Studies. Front. Med. 2020, 7, 598652. [Google Scholar] [CrossRef]
- Shiina, T.; Hosomichi, K.; Inoko, H.; Kulski, J.K. The HLA Genomic Loci Map: Expression, Interaction, Diversity and Disease. J. Hum. Genet. 2009, 54, 15–39. [Google Scholar] [CrossRef]
- Muñoz-Carrillo, J.L.; Castro-García, F.P.; Chávez-Rubalcaba, F.; Chávez-Rubalcaba, I.; Martínez-Rodríguez, J.L.; Hernández-Ruiz, M.E. Immune System Disorders: Hypersensitivity and Autoimmunity. In Immunoregulatory Aspects of Immunotherapy; InTech: Vienna, Austria, 2018. [Google Scholar] [CrossRef]
- Ślebioda, T.J.; Kmieć, Z. Tumour Necrosis Factor Superfamily Members in the Pathogenesis of Inflammatory Bowel Disease. Mediat. Inflamm. 2014, 2014, 325129. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2; Springer New York: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. Inter J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Droździk, M.; Kaczmarek, M.; Malinowski, D.; Broś, U.; Kazienko, A.; Kurzawa, R.; Kurzawski, M. TGFβ3 (TGFB3) Polymorphism Is Associated with Male Infertility. Sci. Rep. 2015, 5, 17151. [Google Scholar] [CrossRef]
- Vendrell, X. New Genetic Point Mutations in Male Infertility. In Reproductomics; Elsevier: Amsterdam, The Netherlands, 2018; pp. 47–62. [Google Scholar] [CrossRef]
- Yin, Y.; Zhu, P.; Luo, T.; Xia, X. Association of Single-nucleotide Polymorphisms in Antioxidant Genes and Their Gene-gene Interactions with Risk of Male Infertility in a Chinese Population. Biomed. Rep. 2020, 13, 49–54. [Google Scholar] [CrossRef]
- van der Ven, K.; Fimmers, R.; Engels, G.; van der Ven, H.; Krebs, D. Evidence for Major Histocompatibility Complex-Mediated Effects on Spermatogenesis in Humans. Hum. Reprod. 2000, 15, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, J.; Zhang, H.; Sun, J.; Sun, Y.; Wang, Z.; Liu, J.; Ding, Q.; Lu, S.; Shi, R.; et al. A Genome-Wide Association Study Reveals That Variants within the HLA Region Are Associated with Risk for Nonobstructive Azoospermia. Am. J. Hum. Genet. 2012, 90, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Stastny, P. HLA-D and IA Antigens in Rheumatoid Arthritis and Systemic Lupus Erythematosus. Arthritis Rheum. 1978, 21, S139–S143. [Google Scholar] [CrossRef]
- Mignot, E.; Hayduk, R.; Black, J.; Grumet, F.C.; Guilleminault, C. HLA DQB1*0602 Is Associated with Cataplexy in 509 Narcoleptic Patients. Sleep 1997, 20, 1012–1020. [Google Scholar] [PubMed]
- Hamza, T.H.; Zabetian, C.P.; Tenesa, A.; Laederach, A.; Montimurro, J.; Yearout, D.; Kay, D.M.; Doheny, K.F.; Paschall, J.; Pugh, E.; et al. Common Genetic Variation in the HLA Region Is Associated with Late-Onset Sporadic Parkinson’s Disease. Nat. Genet. 2010, 42, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Kelly, F.; Alvarez-Lafuente, R.; Alcina, A.; Abad-Grau, M.M.; de las Heras, V.; Lucas, M.; de la Concha, E.G.; Fernández, O.; Arroyo, R.; Matesanz, F.; et al. Members 6B and 14 of the TNF Receptor Superfamily in Multiple Sclerosis Predisposition. Genes Immun. 2011, 12, 145–148. [Google Scholar] [CrossRef]
- Baranzini, S.E.; Khankhanian, P.; Patsopoulos, N.A.; Li, M.; Stankovich, J.; Cotsapas, C.; Søndergaard, H.B.; Ban, M.; Barizzone, N.; Bergamaschi, L.; et al. Network-Based Multiple Sclerosis Pathway Analysis with GWAS Data from 15,000 Cases and 30,000 Controls. Am. J. Hum. Genet. 2013, 92, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Dubois, P.C.A.; Trynka, G.; Franke, L.; Hunt, K.A.; Romanos, J.; Curtotti, A.; Zhernakova, A.; Heap, G.A.R.; Ádány, R.; Aromaa, A.; et al. Multiple Common Variants for Celiac Disease Influencing Immune Gene Expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef]
- Ruyssen-Witrand, A.; Constantin, A.; Cambon-Thomsen, A.; Thomsen, M. New Insights into the Genetics of Immune Responses in Rheumatoid Arthritis. Tissue Antigens 2012, 80, 105–118. [Google Scholar] [CrossRef]
- Cheung, T.C.; Steinberg, M.W.; Oborne, L.M.; Macauley, M.G.; Fukuyama, S.; Sanjo, H.; D’Souza, C.; Norris, P.S.; Pfeffer, K.; Murphy, K.M.; et al. Unconventional Ligand Activation of Herpesvirus Entry Mediator Signals Cell Survival. Proc. Natl. Acad. Sci. USA 2009, 106, 6244–6249. [Google Scholar] [CrossRef]
- Tukey, R.H.; Strassburg, C.P. Human UDP-Glucuronosyltransferases: Metabolism, Expression, and Disease. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 581–616. [Google Scholar] [CrossRef] [PubMed]
- Nandith, P.B.; Adiga, U.; Shenoy, V.; Adiga, M.N.S. UGT1A6 and UGT2B7 Gene Polymorphism and Its Effect in Pediatric Epileptic Patients on Sodium Valproate Monotherapy. Indian J. Pediatrics 2021, 88, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Kua, L.-F.; Ross, S.; Lee, S.-C.; Mimura, K.; Kono, K.; Goh, B.-C.; Yong, W.-P. UGT1A6 Polymorphisms Modulated Lung Cancer Risk in a Chinese Population. PLoS ONE 2012, 7, e42873. [Google Scholar] [CrossRef] [PubMed]
- Justenhoven, C.; Winter, S.; Dünnebier, T.; Hamann, U.; Baisch, C.; Rabstein, S.; Spickenheuer, A.; Harth, V.; Pesch, B.; Brüning, T.; et al. Combined UGT1A1 and UGT1A6 Genotypes Together with a Stressful Life Event Increase Breast Cancer Risk. Breast Cancer Res. Treat. 2010, 124, 289–292. [Google Scholar] [CrossRef]

| Candidate Causal SNP | Functional Class | Gene | Candidate Causal Pathway a | −log10(P) b | In LD with | r2 | D′ | −log10(P) c |
|---|---|---|---|---|---|---|---|---|
| rs8084 | Essential splice site & intronic | HLA-DRA | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 17 24 26 27 32 | - | rs7192 | 0.818 | 1.0 | 5.523 |
| rs7192 | Non synonymous coding | HLA-DRA | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 17 24 26 27 32 | 5.523 | rs7192 | - | - | 5.523 |
| rs7550231 | Regulatory region | TNFRSF14 | 15 29 | - | rs4486391 | 0.96 | 1.0 | 1.867 |
| rs2234167 | Non synonymous coding | TNFRSF14 | 15 29 | - | rs10910093 | 0.89 | 1.0 | 1.745 |
| rs1105879 | Non synonymous coding | UGT1A6 | 18 19 20 21 22 23 25 28 30 31 | - | rs2070959 | 0.825 | 1.0 | 47.000 |
| rs2070959 | Non synonymous coding | UGT1A6 | 18 19 20 21 22 23 25 28 30 31 | 47.000 | rs2070959 | - | - | 47.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvi, R.; Gawde, U.; Idicula-Thomas, S.; Biswas, B. Pathway Analysis of Genome Wide Association Studies (GWAS) Data Associated with Male Infertility. Reprod. Med. 2022, 3, 235-245. https://doi.org/10.3390/reprodmed3030018
Salvi R, Gawde U, Idicula-Thomas S, Biswas B. Pathway Analysis of Genome Wide Association Studies (GWAS) Data Associated with Male Infertility. Reproductive Medicine. 2022; 3(3):235-245. https://doi.org/10.3390/reprodmed3030018
Chicago/Turabian StyleSalvi, Rupashree, Ulka Gawde, Susan Idicula-Thomas, and Barnali Biswas. 2022. "Pathway Analysis of Genome Wide Association Studies (GWAS) Data Associated with Male Infertility" Reproductive Medicine 3, no. 3: 235-245. https://doi.org/10.3390/reprodmed3030018
APA StyleSalvi, R., Gawde, U., Idicula-Thomas, S., & Biswas, B. (2022). Pathway Analysis of Genome Wide Association Studies (GWAS) Data Associated with Male Infertility. Reproductive Medicine, 3(3), 235-245. https://doi.org/10.3390/reprodmed3030018






